Local Population Structure and Patterns of Western Hemisphere Dispersal for Coccidioides spp., the Fungal Cause of Valley Fever
- PMID: 27118594
- PMCID: PMC4850269
- DOI: 10.1128/mBio.00550-16
Local Population Structure and Patterns of Western Hemisphere Dispersal for Coccidioides spp., the Fungal Cause of Valley Fever
Abstract
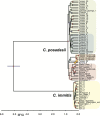
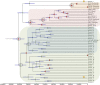
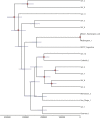
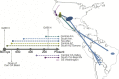
Coccidioidomycosis (or valley fever) is a fungal disease with high morbidity and mortality that affects tens of thousands of people each year. This infection is caused by two sibling species, Coccidioides immitis and C. posadasii, which are endemic to specific arid locales throughout the Western Hemisphere, particularly the desert southwest of the United States. Recent epidemiological and population genetic data suggest that the geographic range of coccidioidomycosis is expanding, as new endemic clusters have been identified in the state of Washington, well outside the established endemic range. The genetic mechanisms and epidemiological consequences of this expansion are unknown and require better understanding of the population structure and evolutionary history of these pathogens. Here we performed multiple phylogenetic inference and population genomics analyses of 68 new and 18 previously published genomes. The results provide evidence of substantial population structure in C. posadasii and demonstrate the presence of distinct geographic clades in central and southern Arizona as well as dispersed populations in Texas, Mexico, South America, and Central America. Although a smaller number of C. immitis strains were included in the analyses, some evidence of phylogeographic structure was also detected in this species, which has been historically limited to California and Baja, Mexico. Bayesian analyses indicated that C. posadasii is the more ancient of the two species and that Arizona contains the most diverse subpopulations. We propose a southern Arizona-northern Mexico origin for C. posadasii and describe a pathway for dispersal and distribution out of this region.
Importance: Coccidioidomycosis, or valley fever, is caused by the pathogenic fungi Coccidioides posadasii and C. immitis The fungal species and disease are primarily found in the American desert southwest, with spotted distribution throughout the Western Hemisphere. Initial molecular studies suggested a likely anthropogenic movement of C. posadasii from North America to South America. Here we comparatively analyze eighty-six genomes of the two Coccidioides species and establish local and species-wide population structures to not only clarify the earlier dispersal hypothesis but also provide evidence of likely ancestral populations and patterns of dispersal for the known subpopulations of C. posadasii.
Copyright © 2016 Engelthaler et al.
Figures


Similar articles
-
Population Structure and Genetic Diversity among Isolates of Coccidioides posadasii in Venezuela and Surrounding Regions.mBio. 2019 Nov 26;10(6):e01976-19. doi: 10.1128/mBio.01976-19. mBio. 2019. PMID: 31772050 Free PMC article.
-
Use of Population Genetics to Assess the Ecology, Evolution, and Population Structure of Coccidioides.Emerg Infect Dis. 2016 Jun;22(6):1022-30. doi: 10.3201/eid2206.151565. Emerg Infect Dis. 2016. PMID: 27191589 Free PMC article.
-
The mysterious desert dwellers: Coccidioides immitis and Coccidioides posadasii, causative fungal agents of coccidioidomycosis.Virulence. 2019 Dec;10(1):222-233. doi: 10.1080/21505594.2019.1589363. Virulence. 2019. PMID: 30898028 Free PMC article. Review.
-
AFLP analysis reveals high genetic diversity but low population structure in Coccidioides posadasii isolates from Mexico and Argentina.BMC Infect Dis. 2013 Sep 3;13:411. doi: 10.1186/1471-2334-13-411. BMC Infect Dis. 2013. PMID: 24004977 Free PMC article.
-
Valley fever under a changing climate in the United States.Environ Int. 2024 Nov;193:109066. doi: 10.1016/j.envint.2024.109066. Epub 2024 Oct 11. Environ Int. 2024. PMID: 39432997 Review.
Cited by
-
Mitochondrial genomes of the human pathogens Coccidioides immitis and Coccidioides posadasii.G3 (Bethesda). 2021 Jul 14;11(7):jkab132. doi: 10.1093/g3journal/jkab132. G3 (Bethesda). 2021. PMID: 33871031 Free PMC article.
-
The Consequences of Our Changing Environment on Life Threatening and Debilitating Fungal Diseases in Humans.J Fungi (Basel). 2021 May 7;7(5):367. doi: 10.3390/jof7050367. J Fungi (Basel). 2021. PMID: 34067211 Free PMC article. Review.
-
Peritoneal and genital coccidioidomycosis in an otherwise healthy Danish female: a case report.BMC Infect Dis. 2017 Jan 31;17(1):105. doi: 10.1186/s12879-017-2212-4. BMC Infect Dis. 2017. PMID: 28143444 Free PMC article.
-
From soil to clinic: current advances in understanding Coccidioides and coccidioidomycosis.Microbiol Mol Biol Rev. 2024 Dec 18;88(4):e0016123. doi: 10.1128/mmbr.00161-23. Epub 2024 Oct 4. Microbiol Mol Biol Rev. 2024. PMID: 39365073 Review.
-
A Recombinant Multivalent Vaccine (rCpa1) Induces Protection for C57BL/6 and HLA Transgenic Mice against Pulmonary Infection with Both Species of Coccidioides.Vaccines (Basel). 2024 Jan 9;12(1):67. doi: 10.3390/vaccines12010067. Vaccines (Basel). 2024. PMID: 38250880 Free PMC article.
References
-
- Marsden-Haug N, Hill H, Litvintseva AP, Engelthaler DM, Driebe EM, Roe CC, Ralston C, Hurst S, Goldoft M, Gade L, Wohrle R, Thompson GR, Brandt ME, Chiller T, Centers for Disease Control and Prevention (CDC) . 2014. Coccidioides immitis identified in soil outside of its known range—Washington, 2013. MMWR Morb Mortal Wkly Rep 63:450. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

