CIB1: a small protein with big ambitions
- PMID: 27118676
- PMCID: PMC4970603
- DOI: 10.1096/fj.201500073R
CIB1: a small protein with big ambitions
Abstract
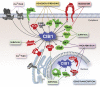
Calcium- and integrin-binding protein 1 (CIB1) is a small, ubiquitously expressed protein that was first identified as an intracellular binding partner of a platelet-specific α-integrin cytoplasmic tail. Although early studies revealed a role for CIB1 in regulating platelet integrin activity, recent studies have indicated a more diverse role for CIB1 in many different cell types and processes, including calcium signaling, migration, adhesion, proliferation, and survival. Increasing evidence also points to a novel role for CIB1 in cancer and cardiovascular disease. In addition, an array of CIB1 binding partners has been identified that provide important insight into how CIB1 may regulate these processes. Some of these binding partners include the serine/threonine kinases, p21-activated kinase 1 (PAK1), apoptosis signal-regulating kinase 1 (ASK1), and polo-like kinase 3 (PLK3). Structural and mutational studies indicate that CIB1 binds most or all of its partners via a well-defined hydrophobic cleft. Although CIB1 itself lacks known enzymatic activity, it supports the PI3K/AKT and MEK/ERK oncogenic signaling pathways, in part, by directly modulating enzymes in these pathways. In this review, we discuss our current understanding of CIB1 and key questions regarding structure and function and how this seemingly diminutive protein impacts important signaling pathways and cellular processes in human health and disease.-Leisner, T. M., Freeman, T. C., Black, J. L., Parise, L. V. CIB1: a small protein with big ambitions.
Keywords: cancer; cardiovascular; kinase; signaling; structure.
© FASEB.
Figures

References
-
- Naik U. P., Patel P. M., Parise L. V. (1997) Identification of a novel calcium-binding protein that interacts with the integrin alphaIIb cytoplasmic domain. J. Biol. Chem. 272, 4651–4654 - PubMed
-
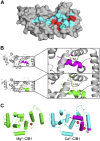
- Gentry H. R., Singer A. U., Betts L., Yang C., Ferrara J. D., Sondek J., Parise L. V. (2005) Structural and biochemical characterization of CIB1 delineates a new family of EF-hand-containing proteins. J. Biol. Chem. 280, 8407–8415 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

