The utility of flow sorting to identify chromosomes carrying a single copy transgene in wheat
- PMID: 27118986
- PMCID: PMC4845436
- DOI: 10.1186/s13007-016-0124-8
The utility of flow sorting to identify chromosomes carrying a single copy transgene in wheat
Abstract
Background: Identification of transgene insertion sites in plant genomes has practical implications for crop breeding and is a stepping stone to analyze transgene function. However, single copy sequences are not always easy to localize in large plant genomes by standard approaches.
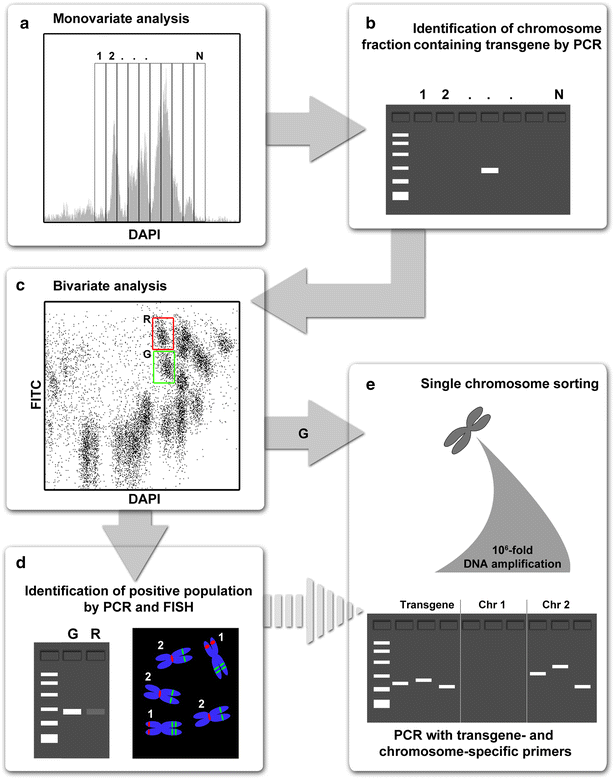
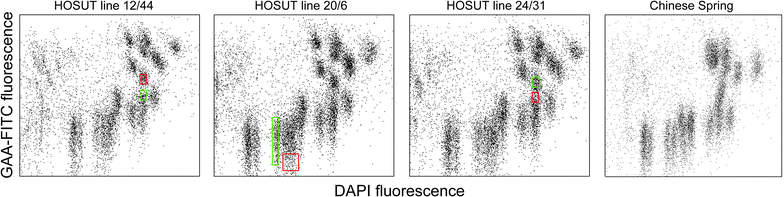
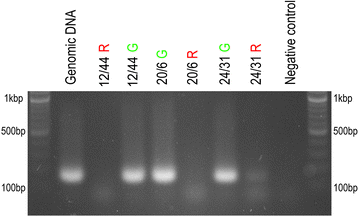
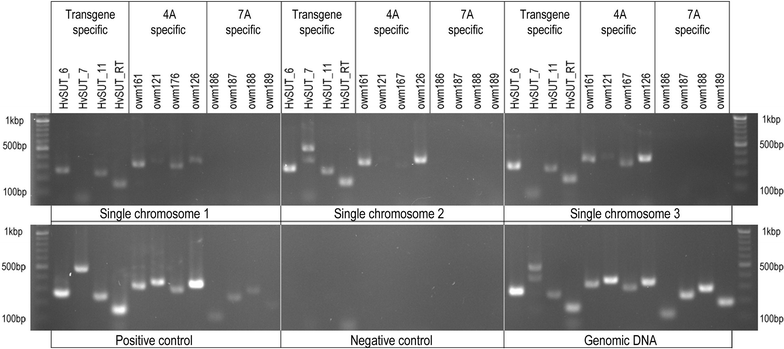
Results: We employed flow cytometric chromosome sorting to determine chromosomal location of barley sucrose transporter construct in three transgenic lines of common wheat. Flow-sorted chromosomes were used as template for PCR and fluorescence in situ hybridization to identify chromosomes with transgenes. The chromosomes carrying the transgenes were then confirmed by PCR using DNA amplified from single flow-sorted chromosomes as template.
Conclusions: Insertion sites of the transgene were unambiguously localized to chromosomes 4A, 7A and 5D in three wheat transgenic lines. The procedure presented in this study is applicable for localization of any single-copy sequence not only in wheat, but in any plant species where suspension of intact mitotic chromosomes suitable for flow cytometric sorting can be prepared.
Keywords: Flow cytometric sorting; Hordeum vulgare; HvSUT1; Single chromosome amplification; Transgene localization; Triticum aestivum.
Figures

References
-
- Fischer RA. The importance of grain or kernel number in wheat: a reply to Sinclair and Jamieson. Field Crops Res. 2008;105:15–21. doi: 10.1016/j.fcr.2007.04.002. - DOI
-
- Svitashev S, Anaiev E, Pawlowski WP, Somers DA. Association of transgene integration sites with chromosome rearrangements in hexaploid oat. Theor Appl Genet. 2000;100:872–880. doi: 10.1007/s001220051364. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

