Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma
- PMID: 27119352
- PMCID: PMC5095006
- DOI: 10.18632/oncotarget.8917
Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma
Abstract
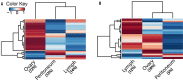
Recently, a class of endogenous species of RNA called circular RNA (circRNA) has been shown to regulate gene expression in mammals and their role in cellular function is just beginning to be understood. To investigate the role of circRNAs in ovarian cancer, we performed paired-end RNA sequencing of primary sites, peritoneal and lymph node metastases from three patients with stage IIIC ovarian cancer. We developed an in-house computational pipeline to identify and characterize the circRNA expression from paired-end RNA-Seq libraries. This pipeline revealed thousands of circular isoforms in Epithelial Ovarian Carcinoma (EOC). These circRNAs are enriched for potentially effective miRNA seed matches. A significantly larger number of circRNAs are differentially expressed between tumor sites than mRNAs. Circular and linear expression exhibits an inverse trend for many cancer related pathways and signaling pathways like NFkB, PI3k/AKT and TGF-β typically activated for mRNA in metastases are inhibited for circRNA expression. Further, circRNAs show a more robust expression pattern across patients than mRNA forms indicating their suitability as biomarkers in highly heterogeneous cancer transcriptomes. The consistency of circular RNA expression may offer new candidates for cancer treatment and prognosis.
Keywords: circular RNA; lymph node metastasis; miRNA; ovarian cancer; peritoneal metastasis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- GLOBOCAN 2012 [ http://globocan.iarc.fr/Default.aspx]
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics. CA: a cancer journal for clinicians. 2009;59:225–49. - PubMed
-
- Morice P, Joulie F, Camatte S, Atallah D, Rouzier R, Pautier P, Pomel C, Lhommé C, Duvillard P, Castaigne D. Lymph node involvement in epithelial ovarian cancer: analysis of 276 pelvic and paraaortic lymphadenectomies and surgical implications. Journal of the American College of Surgeons. 2003;197:198–205. - PubMed
-
- Cannistra SA. Cancer of the ovary. The New England journal of medicine. 2004;351:2519–29. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

