Chemical-Induced Read-Through at Premature Termination Codons Determined by a Rapid Dual-Fluorescence System Based on S. cerevisiae
- PMID: 27119736
- PMCID: PMC4847774
- DOI: 10.1371/journal.pone.0154260
Chemical-Induced Read-Through at Premature Termination Codons Determined by a Rapid Dual-Fluorescence System Based on S. cerevisiae
Abstract
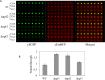
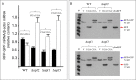
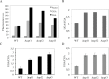
Nonsense mutations generate in-frame stop codons in mRNA leading to a premature arrest of translation. Functional consequences of premature termination codons (PTCs) include the synthesis of truncated proteins with loss of protein function causing severe inherited or acquired diseases. A therapeutic approach has been recently developed that is based on the use of chemical agents with the ability to suppress PTCs (read-through) restoring the synthesis of a functional full-length protein. Research interest for compounds able to induce read-through requires an efficient high throughput large scale screening system. We present a rapid, sensitive and quantitative method based on a dual-fluorescence reporter expressed in the yeast Saccharomyces cerevisiae to monitor and quantitate read-through at PTCs. We have shown that our novel system works equally well in detecting read-through at all three PTCs UGA, UAG and UAA.
Conflict of interest statement
Figures

Similar articles
-
A flow cytometry-based reporter assay identifies macrolide antibiotics as nonsense mutation read-through agents.J Mol Med (Berl). 2016 Apr;94(4):469-82. doi: 10.1007/s00109-015-1364-1. Epub 2015 Dec 1. J Mol Med (Berl). 2016. PMID: 26620677
-
Tobramycin is a suppressor of premature termination codons.J Cyst Fibros. 2013 Dec;12(6):806-11. doi: 10.1016/j.jcf.2013.02.007. Epub 2013 Mar 27. J Cyst Fibros. 2013. PMID: 23540394
-
An homologous in vitro assay for yeast nonsense suppressors.J Biol Chem. 1981 Jul 25;256(14):7298-304. J Biol Chem. 1981. PMID: 7019207
-
Translational readthrough potential of natural termination codons in eucaryotes--The impact of RNA sequence.RNA Biol. 2015;12(9):950-8. doi: 10.1080/15476286.2015.1068497. RNA Biol. 2015. PMID: 26176195 Free PMC article. Review.
-
Targeting Nonsense Mutations in Diseases with Translational Read-Through-Inducing Drugs (TRIDs).BioDrugs. 2016 Apr;30(2):49-74. doi: 10.1007/s40259-016-0157-6. BioDrugs. 2016. PMID: 26886021 Review.
Cited by
-
A genome-wide screen identifies genes that suppress the accumulation of spontaneous mutations in young and aged yeast cells.Aging Cell. 2020 Feb;19(2):e13084. doi: 10.1111/acel.13084. Epub 2019 Dec 18. Aging Cell. 2020. PMID: 31854076 Free PMC article.
-
Innovative Therapies for Cystic Fibrosis: The Road from Treatment to Cure.Mol Diagn Ther. 2019 Apr;23(2):263-279. doi: 10.1007/s40291-018-0372-6. Mol Diagn Ther. 2019. PMID: 30478715 Review.
-
CTELS: A Cell-Free System for the Analysis of Translation Termination Rate.Biomolecules. 2020 Jun 16;10(6):911. doi: 10.3390/biom10060911. Biomolecules. 2020. PMID: 32560154 Free PMC article.
-
Screening Readthrough Compounds to Suppress Nonsense Mutations: Possible Application to β-Thalassemia.J Clin Med. 2020 Jan 21;9(2):289. doi: 10.3390/jcm9020289. J Clin Med. 2020. PMID: 31972957 Free PMC article. Review.
-
High-throughput replica-pinning approach to screen for yeast genes controlling low-frequency events.STAR Protoc. 2022 Jan 13;3(1):101082. doi: 10.1016/j.xpro.2021.101082. eCollection 2022 Mar 18. STAR Protoc. 2022. PMID: 35059655 Free PMC article.
References
-
- Lee HL, Dougherty JP. Pharmaceutical therapies to recode nonsense mutations in inherited diseases. Pharmacology & therapeutics. 2012;136(2):227–66. Epub 2012/07/24. - PubMed
-
- Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nature genetics. 2004;36(8):801–8. Epub 2004/07/31. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

