Lipid dynamics in zebrafish embryonic development observed by DESI-MS imaging and nanoelectrospray-MS
- PMID: 27120110
- PMCID: PMC4916021
- DOI: 10.1039/c6mb00168h
Lipid dynamics in zebrafish embryonic development observed by DESI-MS imaging and nanoelectrospray-MS
Abstract
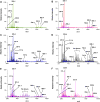
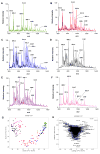
The zebrafish Danio rerio is a model vertebrate organism for understanding biological mechanisms. Recent studies have explored using zebrafish as a model for lipid-related diseases, for in vivo fish bioassays, and for embryonic toxicity experiments. Mass spectrometry (MS) and MS imaging are established tools for lipid profiling and spatial mapping of biomolecules and offer rapid, sensitive, and simple analytical protocols for zebrafish analysis. When ambient ionization techniques are used, ions are generated in native environmental conditions, requiring neither sample preparation nor separation of molecules prior to MS. We used two direct MS techniques to describe the dynamics of the lipid profile during zebrafish embryonic development from 0 to 96 hours post-fertilization and to explore these analytical approaches as molecular diagnostic assays. Desorption electrospray ionization (DESI) MS imaging followed by nanoelectrospray (nESI) MS and tandem MS (MS/MS) were used in positive and negative ion modes, allowing the detection of a large variety of phosphatidylglycerols, phosphatidylcholines, phosphatidylinositols, free fatty acids, triacylglycerols, ubiquinone, squalene, and other lipids, and revealed information on the spatial distributions of lipids within the embryo and on lipid molecular structure. Differences were observed in the relative ion abundances of free fatty acids, triacylglycerols, and ubiquinone - essentially localized to the yolk - across developmental stages, whereas no relevant differences were found in the distribution of complex membrane glycerophospholipids, indicating conserved lipid constitution. Embryos exposed to trichloroethylene for 72 hours exhibited an altered lipid profile, indicating the potential utility of this technique for testing the effects of environmental contaminants.
Conflict of interest statement
No conflicts of interest are declared.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

