Evolved plasmid-host interactions reduce plasmid interference cost
- PMID: 27121483
- PMCID: PMC5024541
- DOI: 10.1111/mmi.13407
Evolved plasmid-host interactions reduce plasmid interference cost
Abstract
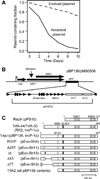
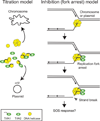
Antibiotic selection drives adaptation of antibiotic resistance plasmids to new bacterial hosts, but the molecular mechanisms are still poorly understood. We previously showed that a broad-host-range plasmid was poorly maintained in Shewanella oneidensis, but rapidly adapted through mutations in the replication initiation gene trfA1. Here we examined if these mutations reduced the fitness cost of TrfA1, and whether this was due to changes in interaction with the host's DNA helicase DnaB. The strains expressing evolved TrfA1 variants showed a higher growth rate than those expressing ancestral TrfA1. The evolved TrfA1 variants showed a lower affinity to the helicase than ancestral TrfA1 and were no longer able to activate the helicase at the oriV without host DnaA. Moreover, persistence of the ancestral plasmid was increased upon overexpression of DnaB. Finally, the evolved TrfA1 variants generated higher plasmid copy numbers than ancestral TrfA1. The findings suggest that ancestral plasmid instability can at least partly be explained by titration of DnaB by TrfA1. Thus under antibiotic selection resistance plasmids can adapt to a novel bacterial host through partial loss of function mutations that simultaneously increase plasmid copy number and decrease unfavorably high affinity to one of the hosts' essential proteins.
© 2016 The Authors. Molecular Microbiology Published by John Wiley & Sons Ltd.
Conflict of interest statement
None of the authors have a conflict of interest.
Figures





Similar articles
-
The role of clonal interference in the evolutionary dynamics of plasmid-host adaptation.mBio. 2012 Jul 3;3(4):e00077-12. doi: 10.1128/mBio.00077-12. Print 2012. mBio. 2012. PMID: 22761390 Free PMC article.
-
Roles of long and short replication initiation proteins in the fate of IncP-1 plasmids.J Bacteriol. 2012 Mar;194(6):1533-43. doi: 10.1128/JB.06395-11. Epub 2012 Jan 6. J Bacteriol. 2012. PMID: 22228734 Free PMC article.
-
A multifunctional plasmid-encoded replication initiation protein both recruits and positions an active helicase at the replication origin.Proc Natl Acad Sci U S A. 2003 Jul 22;100(15):8692-7. doi: 10.1073/pnas.1532393100. Epub 2003 Jun 30. Proc Natl Acad Sci U S A. 2003. PMID: 12835421 Free PMC article.
-
Strategies for helicase recruitment and loading in bacteria.EMBO Rep. 2003 Jan;4(1):37-41. doi: 10.1038/sj.embor.embor703. EMBO Rep. 2003. PMID: 12524518 Free PMC article. Review.
-
Mechanisms of Theta Plasmid Replication in Enterobacteria and Implications for Adaptation to Its Host.EcoSal Plus. 2020 Nov;9(1):10.1128/ecosalplus.ESP-0026-2019. doi: 10.1128/ecosalplus.ESP-0026-2019. EcoSal Plus. 2020. PMID: 33210586 Free PMC article. Review.
Cited by
-
Genomics, Transcriptomics, and Metabolomics Reveal That Minimal Modifications in the Host Are Crucial for the Compensatory Evolution of ColE1-Like Plasmids.mSphere. 2022 Dec 21;7(6):e0018422. doi: 10.1128/msphere.00184-22. Epub 2022 Nov 23. mSphere. 2022. PMID: 36416553 Free PMC article.
-
Plasmid stability is enhanced by higher-frequency pulses of positive selection.Proc Biol Sci. 2018 Jan 10;285(1870):20172497. doi: 10.1098/rspb.2017.2497. Proc Biol Sci. 2018. PMID: 29321301 Free PMC article.
-
Plasmid Viability Depends on the Ecological Setting of Hosts within a Multiplasmid Community.Microbiol Spectr. 2022 Apr 27;10(2):e0013322. doi: 10.1128/spectrum.00133-22. Epub 2022 Apr 13. Microbiol Spectr. 2022. PMID: 35416702 Free PMC article.
-
Persistence of plasmid and tet(X4) in an Escherichia coli isolate coharboring bla NDM-5 and mcr-1 after acquiring an IncFII tet(X4)-positive plasmid.Front Microbiol. 2022 Oct 19;13:1010387. doi: 10.3389/fmicb.2022.1010387. eCollection 2022. Front Microbiol. 2022. PMID: 36338060 Free PMC article.
-
Emerging patterns of plasmid-host coevolution that stabilize antibiotic resistance.Sci Rep. 2017 Jul 7;7(1):4853. doi: 10.1038/s41598-017-04662-0. Sci Rep. 2017. PMID: 28687759 Free PMC article.
References
-
- Bagdasarian M, Lurz R, Ruckert B, Franklin FC, Bagdasarian MM, Frey J, Timmis KN. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in. Pseudomonas. Gene. 1981;16:237–247. - PubMed
-
- Bouma JE, Lenski RE. Evolution of a bacteria/plasmid association. Nature. 1988;335:351–352. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

