CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq
- PMID: 27121950
- PMCID: PMC4848782
- DOI: 10.1186/s13059-016-0938-8
CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq
Abstract
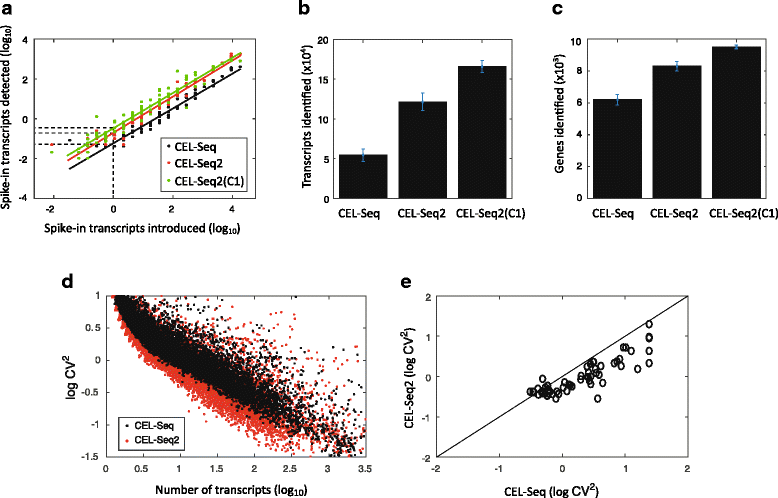
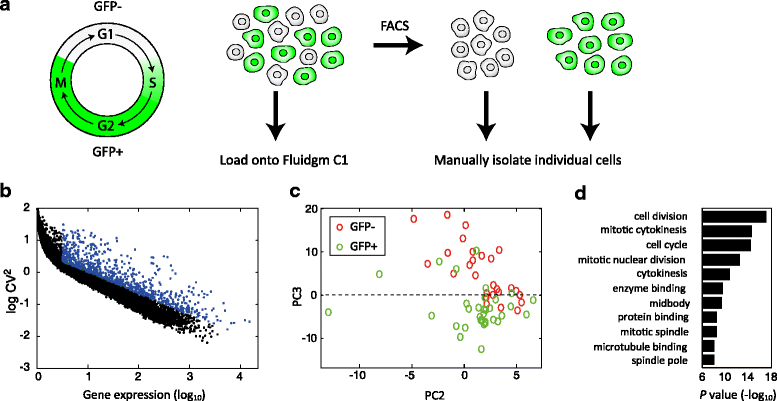
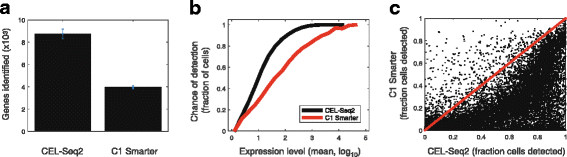
Single-cell transcriptomics requires a method that is sensitive, accurate, and reproducible. Here, we present CEL-Seq2, a modified version of our CEL-Seq method, with threefold higher sensitivity, lower costs, and less hands-on time. We implemented CEL-Seq2 on Fluidigm's C1 system, providing its first single-cell, on-chip barcoding method, and we detected gene expression changes accompanying the progression through the cell cycle in mouse fibroblast cells. We also compare with Smart-Seq to demonstrate CEL-Seq2's increased sensitivity relative to other available methods. Collectively, the improvements make CEL-Seq2 uniquely suited to single-cell RNA-Seq analysis in terms of economics, resolution, and ease of use.
Figures

References
-
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–82. - PubMed
-
- Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques. 2001;30:892–7. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

