Terrace Aware Data Structure for Phylogenomic Inference from Supermatrices
- PMID: 27121966
- PMCID: PMC5066062
- DOI: 10.1093/sysbio/syw037
Terrace Aware Data Structure for Phylogenomic Inference from Supermatrices
Abstract
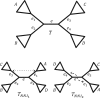
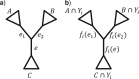
In phylogenomics the analysis of concatenated gene alignments, the so-called supermatrix, is commonly accompanied by the assumption of partition models. Under such models each gene, or more generally partition, is allowed to evolve under its own evolutionary model. Although partition models provide a more comprehensive analysis of supermatrices, missing data may hamper the tree search algorithms due to the existence of phylogenetic (partial) terraces. Here, we introduce the phylogenetic terrace aware (PTA) data structure for the efficient analysis under partition models. In the presence of missing data PTA exploits (partial) terraces and induced partition trees to save computation time. We show that an implementation of PTA in IQ-TREE leads to a substantial speedup of up to 4.5 and 8 times compared with the standard IQ-TREE and RAxML implementations, respectively. PTA is generally applicable to all types of partition models and common topological rearrangements thus can be employed by all phylogenomic inference software.
Keywords: Maximum likelihood; partial terraces; partition models; phylogenetic terraces; phylogenomic inference.
© The Author(s) 2016. Published by Oxford University Press, on behalf of the Society of Systematic Biologists.
Figures




References
-
- Bininda-Emonds O.R., Gittleman J.L., Purvis A. 1999. Building large trees by combining phylogenetic information: a complete phylogeny of the extant Carnivora (Mammalia). Biol. Rev. Camb. Philos. Soc. 74(2):143–175. - PubMed
-
- Bininda-Emonds O.R.P., Gittleman J.L., Steel M.A. 2002. The (Super)tree of life: Procedures, problems, and prospects. Annu. Rev. Ecol. Syst. 33:265–289.
-
- Bouchenak-Khelladi Y., Salamin N., Savolainen V., Forest F., Bank M.V., Chase M.W., Hodkinson T.R. 2008. Large multi-gene phylogenetic trees of the grasses (Poaceae): Progress towards complete tribal and generic level sampling. Mol. Phylogenet. Evol. 47(2):488–505. - PubMed
-
- De Queiroz A., Donoghue M.J., Kim J. 1995. Separate versus combined analysis of phylogenetic evidence. Annu. Rev. Ecol. Syst. 26:657–681.
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

