Mutations in Cockayne Syndrome-Associated Genes (Csa and Csb) Predispose to Cisplatin-Induced Hearing Loss in Mice
- PMID: 27122034
- PMCID: PMC4846672
- DOI: 10.1523/JNEUROSCI.3890-15.2016
Mutations in Cockayne Syndrome-Associated Genes (Csa and Csb) Predispose to Cisplatin-Induced Hearing Loss in Mice
Abstract
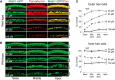
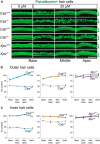
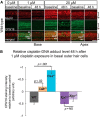
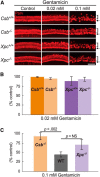
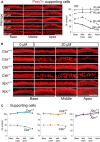
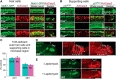
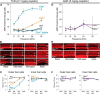
Cisplatin is a common and effective chemotherapeutic agent, yet it often causes permanent hearing loss as a result of sensory hair cell death. The causes of sensitivity to DNA-damaging agents in nondividing cell populations, such as cochlear hair and supporting cells, are poorly understood, as are the specific DNA repair pathways that protect these cells. Nucleotide excision repair (NER) is a conserved and versatile DNA repair pathway for many DNA-distorting lesions, including cisplatin-DNA adducts. Progressive sensorineural hearing loss is observed in a subset of NER-associated DNA repair disorders including Cockayne syndrome and some forms of xeroderma pigmentosum. We investigated whether either of the two overlapping branches that encompass NER, transcription-coupled repair or global genome repair, which are implicated in Cockayne syndrome and xeroderma pigmentosum group C, respectively, modulates cisplatin-induced hearing loss and cell death in the organ of Corti, the auditory sensory epithelium of mammals. We report that cochlear hair cells and supporting cells in transcription-coupled repair-deficient Cockayne syndrome group A (Csa(-/-)) and group B (Csb(-/-)) mice are hypersensitive to cisplatin, in contrast to global genome repair-deficient Xpc(-/-) mice, both in vitro and in vivo We show that sensory hair cells in Csa(-/-) and Csb(-/-) mice fail to remove cisplatin-DNA adducts efficiently in vitro; and unlike Xpc(-/-) mice, Csa(-/-) and Csb(-/-) mice lose hearing and manifest outer hair cell degeneration after systemic cisplatin treatment. Our results demonstrate that Csa and Csb deficiencies predispose to cisplatin-induced hearing loss and hair/supporting cell damage in the mammalian organ of Corti, and emphasize the importance of transcription-coupled DNA repair in the protection against cisplatin ototoxicity.
Significance statement: The utility of cisplatin in chemotherapy remains limited due to serious side effects, including sensorineural hearing loss. We show that mouse models of Cockayne syndrome, a progeroid disorder resulting from a defect in the transcription-coupled DNA repair (TCR) branch of nucleotide excision repair, are hypersensitive to cisplatin-induced hearing loss and sensory hair cell death in the organ of Corti, the mammalian auditory sensory epithelium. Our work indicates that Csa and Csb, two genes involved in TCR, are preferentially required to protect against cisplatin ototoxicity, relative to global genome repair-specific elements of nucleotide excision repair, and suggests that TCR is a major force maintaining DNA integrity in the cochlea. The Cockayne syndrome mice thus represent a model for testing the contribution of DNA repair mechanisms to cisplatin ototoxicity.
Keywords: Cockayne syndrome; DNA repair; cisplatin; hearing loss; ototoxicity; sensory hair cells.
Copyright © 2016 the authors 0270-6474/16/364758-13$15.00/0.
Figures
Similar articles
-
Cockayne syndrome group B (Csb) and group a (Csa) deficiencies predispose to hearing loss and cochlear hair cell degeneration in mice.J Neurosci. 2015 Mar 11;35(10):4280-6. doi: 10.1523/JNEUROSCI.5063-14.2015. J Neurosci. 2015. PMID: 25762674 Free PMC article.
-
Age-related neuronal degeneration: complementary roles of nucleotide excision repair and transcription-coupled repair in preventing neuropathology.PLoS Genet. 2011 Dec;7(12):e1002405. doi: 10.1371/journal.pgen.1002405. Epub 2011 Dec 8. PLoS Genet. 2011. PMID: 22174697 Free PMC article.
-
The relationship between benzo[a]pyrene-induced mutagenesis and carcinogenesis in repair-deficient Cockayne syndrome group B mice.Cancer Res. 2000 Oct 15;60(20):5681-7. Cancer Res. 2000. PMID: 11059760
-
Bacterial DNA repair genes and their eukaryotic homologues: 4. The role of nucleotide excision DNA repair (NER) system in mammalian cells.Acta Biochim Pol. 2007;54(3):469-82. Epub 2007 Sep 23. Acta Biochim Pol. 2007. PMID: 17893751 Review.
-
Cockayne syndrome group B (CSB) protein: at the crossroads of transcriptional networks.Mech Ageing Dev. 2013 May-Jun;134(5-6):234-42. doi: 10.1016/j.mad.2013.03.004. Epub 2013 Apr 3. Mech Ageing Dev. 2013. PMID: 23562425 Review.
Cited by
-
Comparative study of cytotoxic effects induced by environmental genotoxins using XPC- and CSB-deficient human lymphoblastoid TK6 cells.Genes Environ. 2019 Jul 16;41:15. doi: 10.1186/s41021-019-0130-y. eCollection 2019. Genes Environ. 2019. PMID: 31346351 Free PMC article.
-
Insights into platinum-induced peripheral neuropathy-current perspective.Neural Regen Res. 2020 Sep;15(9):1623-1630. doi: 10.4103/1673-5374.276321. Neural Regen Res. 2020. PMID: 32209761 Free PMC article. Review.
-
The Mechanotransduction Channel and Organic Cation Transporter Are Critical for Cisplatin Ototoxicity in Murine Hair Cells.Front Mol Neurosci. 2022 Feb 10;15:835448. doi: 10.3389/fnmol.2022.835448. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35221917 Free PMC article.
-
CDK2 regulates aminoglycoside-induced hair cell death through modulating c-Jun activity: Inhibiting CDK2 to preserve hearing.Front Mol Neurosci. 2022 Oct 13;15:1013383. doi: 10.3389/fnmol.2022.1013383. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36311033 Free PMC article.
-
Sex Differences in Aging: Genomic Instability.J Gerontol A Biol Sci Med Sci. 2018 Jan 16;73(2):166-174. doi: 10.1093/gerona/glx105. J Gerontol A Biol Sci Med Sci. 2018. PMID: 28575157 Free PMC article. Review.
References
-
- Berg RJ, Rebel H, van der Horst GT, van Kranen HJ, Mullenders LH, van Vloten WA, de Gruijl FR. Impact of global genome repair versus transcription-coupled repair on ultraviolet carcinogenesis in hairless mice. Cancer Res. 2000;60:2858–2863. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
