Use of Shotgun Metagenome Sequencing To Detect Fecal Colonization with Multidrug-Resistant Bacteria in Children
- PMID: 27122381
- PMCID: PMC4922122
- DOI: 10.1128/JCM.02638-15
Use of Shotgun Metagenome Sequencing To Detect Fecal Colonization with Multidrug-Resistant Bacteria in Children
Abstract
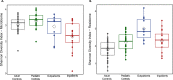
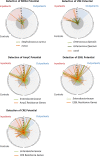
Prevention of multidrug-resistant (MDR) bacterial infections relies on accurate detection of these organisms. We investigated shotgun metagenome sequencing for the detection of methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus (VRE), and MDR Enterobacteriaceae Fecal metagenomes were analyzed from high-risk inpatients and compared to those of low-risk outpatients and controls with minimal risk for a MDR bacterial infection. Principal-component analysis clustered patient samples into distinct cohorts, confirming that the microbiome composition was significantly different between cohorts (P = 0.006). Microbial diversity and relative anaerobe abundance were preserved in outpatients compared to those in controls. Relative anaerobe abundance was significantly reduced in inpatients compared to that in outpatients (P = 0.006). Although the potential for MDR bacteria was increased in inpatients and outpatients compared to that in controls (P < 0.001), there was no difference between inpatients and outpatients. However, 9 (53%) inpatients had colonization with a MDR bacterium that was not identified by culture. Unlike culture, shotgun sequencing quantitatively characterizes the burdens of multiple MDR bacteria relative to all of the microbiota within the intestinal community. We propose consideration of key microbiome features, such as diversity and relative anaerobe abundance, in addition to the detection of MDR bacteria by shotgun metagenome sequencing as a novel method that might better identify patients who are at increased risk of a MDR infection.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Donskey CJ, Chowdhry TK, Hecker MT, Hoyen CK, Hanrahan JA, Hujer AM, Hutton-Thomas RA, Whalen CC, Bonomo RA, Rice LB. 2000. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N Engl J Med 343:1925–1932. doi:10.1056/NEJM200012283432604. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

