Thermostability of Enzymes from Molecular Dynamics Simulations
- PMID: 27123810
- PMCID: PMC4977845
- DOI: 10.1021/acs.jctc.6b00120
Thermostability of Enzymes from Molecular Dynamics Simulations
Abstract
Thermodynamic stability is a central requirement for protein function, and one goal of protein engineering is improvement of stability, particularly for applications in biotechnology. Herein, molecular dynamics simulations are used to predict in vitro thermostability of members of the bacterial ribonuclease HI (RNase H) family of endonucleases. The temperature dependence of the generalized order parameter, S, for four RNase H homologues, from psychrotrophic, mesophilic, and thermophilic organisms, is highly correlated with experimentally determined melting temperatures and with calculated free energies of folding at the midpoint temperature of the simulations. This study provides an approach for in silico mutational screens to improve thermostability of biologically and industrially relevant enzymes.
Figures

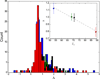
References
-
- Bornscheuer UT, Huisman GW, Kazlauskas RJ, Lutz S, Moore JC, Robins K. Engineering the third wave of biocatalysis. Nature. 2012;485:185–194. - PubMed
-
- Zamost BL, Nielsen HK, Starnes RL. Thermostable enzymes for industrial applications. J. Ind. Microbiol. 1991;8:71–81.
-
- Kristjansson JK. Thermophilic organisms as sources of thermostable enzymes. Trends Biotechnol. 1989;7:349–353.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
