Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity
- PMID: 27126040
- PMCID: PMC5240844
- DOI: 10.1126/science.aad3369
Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity
Abstract
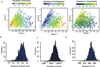
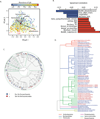
Deep sequencing of the gut microbiomes of 1135 participants from a Dutch population-based cohort shows relations between the microbiome and 126 exogenous and intrinsic host factors, including 31 intrinsic factors, 12 diseases, 19 drug groups, 4 smoking categories, and 60 dietary factors. These factors collectively explain 18.7% of the variation seen in the interindividual distance of microbial composition. We could associate 110 factors to 125 species and observed that fecal chromogranin A (CgA), a protein secreted by enteroendocrine cells, was exclusively associated with 61 microbial species whose abundance collectively accounted for 53% of microbial composition. Low CgA concentrations were seen in individuals with a more diverse microbiome. These results are an important step toward a better understanding of environment-diet-microbe-host interactions.
Copyright © 2016, American Association for the Advancement of Science.
Conflict of interest statement
The authors have no conflicts of interest to report.
Figures


References
-
- Scholtens S, et al. Cohort Profile: LifeLines, a three-generation cohort study and biobank. Int. J. Epidemiol. 2014;44:1172–1180. - PubMed
-
-
Information on materials and methods is available at the Science Web site
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

