Near real-time monitoring of HIV transmission hotspots from routine HIV genotyping: an implementation case study
- PMID: 27126490
- PMCID: PMC4853759
- DOI: 10.1016/S2352-3018(16)00046-1
Near real-time monitoring of HIV transmission hotspots from routine HIV genotyping: an implementation case study
Abstract
Background: HIV evolves rapidly and therefore infections with similar genetic sequences are likely linked by recent transmission events. Clusters of related infections can represent subpopulations with high rates of transmission. We describe the implementation of an automated near real-time system to monitor and characterise HIV transmission hotspots in British Columbia, Canada.
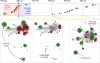
Methods: In this implementation case study, we applied a monitoring system to the British Columbia drug treatment database, which holds more than 32 000 anonymised HIV genotypes for nearly 9000 residents of British Columbia living with HIV. On average, five to six new HIV genotypes are deposited in the database every day, which triggers an automated reanalysis of the entire database. We extracted clusters of five or more individuals with short phylogenetic distances between their respective HIV sequences. The system generated monthly reports of the growth and characteristics of clusters that were distributed to public health officers.
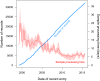
Findings: In June, 2014, the monitoring system detected the expansion of a cluster by 11 new cases during 3 months, including eight cases with transmitted drug resistance. This cluster generally comprised young men who have sex with men. The subsequent report precipitated an enhanced public health follow-up to ensure linkage to care and treatment initiation in the affected subpopulation. Of the nine cases associated with this follow-up, all had already been linked to care and five cases had started treatment. Subsequent to the follow-up, three additional cases started treatment and most cases achieved suppressed viral loads. During the next 12 months, we detected 12 new cases in this cluster with reduction in the onward transmission of drug resistance.
Interpretation: Our findings show the first application of an automated phylogenetic system monitoring a clinical database to detect a recent HIV outbreak and support the ensuing public health response. By making secondary use of routinely collected HIV genotypes, this approach is cost-effective, attains near real-time monitoring of new cases, and can be implemented in all settings in which HIV genotyping is the standard of care.
Funding: BC Centre for Excellence in HIV/AIDS, the Canadian Institutes for Health Research, the Genome Canada-CIHR Partnership in Genomics and Personalized Health, and the US National Institute on Drug Abuse.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Figures

Comment in
-
Translating phylogeny into action for HIV surveillance.Lancet HIV. 2016 May;3(5):e196-7. doi: 10.1016/S2352-3018(16)30012-1. Epub 2016 Apr 7. Lancet HIV. 2016. PMID: 27126483 No abstract available.
-
Need for robust and inclusive public health ethics review of the monitoring of HIV phylogenetic clusters for HIV prevention.Lancet HIV. 2016 Oct;3(10):e461. doi: 10.1016/S2352-3018(16)30156-4. Lancet HIV. 2016. PMID: 27687039 No abstract available.
References
-
- Gunthard HF, Aberg JA, Eron JJ, et al. Antiretroviral treatment of adult HIV infection: 2014 recommendations of the International Antiviral Society-USA Panel. JAMA. 2014;312(4):410–25. - PubMed
-
- Tural C, Ruiz L, Holtzer C, et al. Clinical utility of HIV-1 genotyping and expert advice: the Havana trial. AIDS. 2002;16(2):209–18. - PubMed
-
- Williamson S. Adaptation in the env gene of HIV-1 and evolutionary theories of disease progression. Mol Biol Evol. 2003;20(8):1318–25. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

