Somatic genomic alterations in retinoblastoma beyond RB1 are rare and limited to copy number changes
- PMID: 27126562
- PMCID: PMC4850475
- DOI: 10.1038/srep25264
Somatic genomic alterations in retinoblastoma beyond RB1 are rare and limited to copy number changes
Abstract
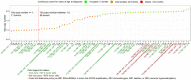
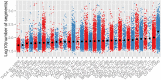
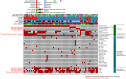
Retinoblastoma is a rare childhood cancer initiated by RB1 mutation or MYCN amplification, while additional alterations may be required for tumor development. However, the view on single nucleotide variants is very limited. To better understand oncogenesis, we determined the genomic landscape of retinoblastoma. We performed exome sequencing of 71 retinoblastomas and matched blood DNA. Next, we determined the presence of single nucleotide variants, copy number alterations and viruses. Aside from RB1, recurrent gene mutations were very rare. Only a limited fraction of tumors showed BCOR (7/71, 10%) or CREBBP alterations (3/71, 4%). No evidence was found for the presence of viruses. Instead, specific somatic copy number alterations were more common, particularly in patients diagnosed at later age. Recurrent alterations of chromosomal arms often involved less than one copy, also in highly pure tumor samples, suggesting within-tumor heterogeneity. Our results show that retinoblastoma is among the least mutated cancers and signify the extreme sensitivity of the childhood retina for RB1 loss. We hypothesize that retinoblastomas arising later in retinal development benefit more from subclonal secondary alterations and therefore, these alterations are more selected for in these tumors. Targeted therapy based on these subclonal events might be insufficient for complete tumor control.
Figures



References
-
- Linabery A. M. & Ross J. A. Trends in childhood cancer incidence in the U.S. (1992–2004). Cancer 112, 416–32 (2008). - PubMed
-
- Parkin D. M., Stiller C. A., Draper G. J. & Bieber C. A. The international incidence of childhood cancer. Int. J. Cancer 42, 511–520 (1988). - PubMed
-
- Rushlow D. E. et al. Characterisation of retinoblastomas without RB1 mutations: genomic, gene expression, and clinical studies. Lancet. Oncol. 14, 327–34 (2013). - PubMed
-
- Dimaras H. et al. Loss of RB1 induces non-proliferative retinoma: increasing genomic instability correlates with progression to retinoblastoma. Hum. Mol. Genet. 17, 1363–1372 (2008). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

