The Ontology for Biomedical Investigations
- PMID: 27128319
- PMCID: PMC4851331
- DOI: 10.1371/journal.pone.0154556
The Ontology for Biomedical Investigations
Abstract
The Ontology for Biomedical Investigations (OBI) is an ontology that provides terms with precisely defined meanings to describe all aspects of how investigations in the biological and medical domains are conducted. OBI re-uses ontologies that provide a representation of biomedical knowledge from the Open Biological and Biomedical Ontologies (OBO) project and adds the ability to describe how this knowledge was derived. We here describe the state of OBI and several applications that are using it, such as adding semantic expressivity to existing databases, building data entry forms, and enabling interoperability between knowledge resources. OBI covers all phases of the investigation process, such as planning, execution and reporting. It represents information and material entities that participate in these processes, as well as roles and functions. Prior to OBI, it was not possible to use a single internally consistent resource that could be applied to multiple types of experiments for these applications. OBI has made this possible by creating terms for entities involved in biological and medical investigations and by importing parts of other biomedical ontologies such as GO, Chemical Entities of Biological Interest (ChEBI) and Phenotype Attribute and Trait Ontology (PATO) without altering their meaning. OBI is being used in a wide range of projects covering genomics, multi-omics, immunology, and catalogs of services. OBI has also spawned other ontologies (Information Artifact Ontology) and methods for importing parts of ontologies (Minimum information to reference an external ontology term (MIREOT)). The OBI project is an open cross-disciplinary collaborative effort, encompassing multiple research communities from around the globe. To date, OBI has created 2366 classes and 40 relations along with textual and formal definitions. The OBI Consortium maintains a web resource (http://obi-ontology.org) providing details on the people, policies, and issues being addressed in association with OBI. The current release of OBI is available at http://purl.obolibrary.org/obo/obi.owl.
Conflict of interest statement
Figures
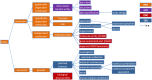
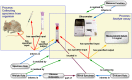
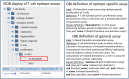
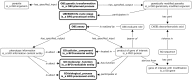

References
Publication types
MeSH terms
Grants and funding
- BB/C008200/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 GM093132/GM/NIGMS NIH HHS/United States
- HHSN272201200010C/AI/NIAID NIH HHS/United States
- U01 DK072473/DK/NIDDK NIH HHS/United States
- BB/I000771/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- U24 DA039832/DA/NIDA NIH HHS/United States
- R24 OD011883/OD/NIH HHS/United States
- HHSN272201400028C/AI/NIAID NIH HHS/United States
- P41 HG003619/HG/NHGRI NIH HHS/United States
- HHSN272201400030C/AI/NIAID NIH HHS/United States
- BB/G000638/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- U19 AI118626/AI/NIAID NIH HHS/United States
- BB/E025080/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 AI081062/AI/NIAID NIH HHS/United States
- R01 GM080646/GM/NIGMS NIH HHS/United States
- UL1 TR001412/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

