Competence Classification of Cumulus and Granulosa Cell Transcriptome in Embryos Matched by Morphology and Female Age
- PMID: 27128483
- PMCID: PMC4851390
- DOI: 10.1371/journal.pone.0153562
Competence Classification of Cumulus and Granulosa Cell Transcriptome in Embryos Matched by Morphology and Female Age
Abstract
Objective: By focussing on differences in the mural granulosa cell (MGC) and cumulus cell (CC) transcriptomes from follicles resulting in competent (live birth) and non-competent (no pregnancy) oocytes the study aims on defining a competence classifier expression profile in the two cellular compartments.
Design: A case-control study.
Setting: University based facilities for clinical services and research.
Patients: MGC and CC samples from 60 women undergoing IVF treatment following the long GnRH-agonist protocol were collected. Samples from 16 oocytes where live birth was achieved and 16 age- and embryo morphology matched incompetent oocytes were included in the study.
Methods: MGC and CC were isolated immediately after oocyte retrieval. From the 16 competent and non-competent follicles, mRNA was extracted and expression profile generated on the Human Gene 1.0 ST Affymetrix array. Live birth prediction analysis using machine learning algorithms (support vector machines) with performance estimation by leave-one-out cross validation and independent validation on an external data set.
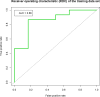
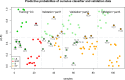
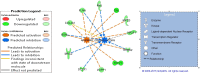
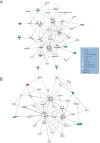
Results: We defined a signature of 30 genes expressed in CC predictive of live birth. This live birth prediction model had an accuracy of 81%, a sensitivity of 0.83, a specificity of 0.80, a positive predictive value of 0.77, and a negative predictive value of 0.86. Receiver operating characteristic analysis found an area under the curve of 0.86, significantly greater than random chance. When applied on 3 external data sets with the end-point outcome measure of blastocyst formation, the signature resulted in 62%, 75% and 88% accuracy, respectively. The genes in the classifier are primarily connected to apoptosis and involvement in formation of extracellular matrix. We were not able to define a robust MGC classifier signature that could classify live birth with accuracy above random chance level.
Conclusion: We have developed a cumulus cell classifier, which showed a promising performance on external data. This suggests that the gene signature at least partly include genes that relates to competence in the developing blastocyst.
Conflict of interest statement
Figures
Similar articles
-
Comparison of gene expression profiles in granulosa and cumulus cells after ovulation induction with either human chorionic gonadotropin or a gonadotropin-releasing hormone agonist trigger.Fertil Steril. 2013 Oct;100(4):994-1001. doi: 10.1016/j.fertnstert.2013.05.038. Epub 2013 Jul 12. Fertil Steril. 2013. PMID: 23856575 Clinical Trial.
-
Ranking and selection of MII oocytes in human ICSI cycles using gene expression levels from associated cumulus cells.Hum Reprod. 2013 Nov;28(11):2930-42. doi: 10.1093/humrep/det357. Epub 2013 Sep 15. Hum Reprod. 2013. PMID: 24041818
-
Differences in cumulus cell gene expression indicate the benefit of a pre-maturation step to improve in-vitro bovine embryo production.Mol Hum Reprod. 2016 Dec;22(12):882-897. doi: 10.1093/molehr/gaw055. Epub 2016 Aug 24. Mol Hum Reprod. 2016. PMID: 27559149
-
The transcriptome of follicular cells: biological insights and clinical implications for the treatment of infertility.Hum Reprod Update. 2014 Jan-Feb;20(1):1-11. doi: 10.1093/humupd/dmt044. Epub 2013 Sep 29. Hum Reprod Update. 2014. PMID: 24082041 Free PMC article. Review.
-
CUMULUS CELL GENES AS POTENTIAL BIOMARKERS OF OOCYTE AND EMBRYO DEVELOPMENTAL COMPETENCE.Fiziol Zh (1994). 2016;62(1):107-13. doi: 10.15407/fz62.01.107. Fiziol Zh (1994). 2016. PMID: 29537212 Review.
Cited by
-
mtDNA content in cumulus cells does not predict development to blastocyst or implantation.Hum Reprod Open. 2022 Jul 6;2022(3):hoac029. doi: 10.1093/hropen/hoac029. eCollection 2022. Hum Reprod Open. 2022. PMID: 35864920 Free PMC article.
-
Brilliant cresyl blue enhanced optoacoustic imaging enables non-destructive imaging of mammalian ovarian follicles for artificial reproduction.J R Soc Interface. 2020 Nov;17(172):20200776. doi: 10.1098/rsif.2020.0776. Epub 2020 Nov 4. J R Soc Interface. 2020. PMID: 33143591 Free PMC article.
-
Maturational competence of equine oocytes is associated with alterations in their 'cumulome'.Mol Hum Reprod. 2024 Sep 12;30(9):gaae033. doi: 10.1093/molehr/gaae033. Mol Hum Reprod. 2024. PMID: 39288330 Free PMC article.
-
Polybrominated Diphenyl Ethers in Human Follicular Fluid Dysregulate Mural and Cumulus Granulosa Cell Gene Expression.Endocrinology. 2021 Mar 1;162(3):bqab003. doi: 10.1210/endocr/bqab003. Endocrinology. 2021. PMID: 33543239 Free PMC article.
-
Comparison of Multivariable Logistic Regression and Other Machine Learning Algorithms for Prognostic Prediction Studies in Pregnancy Care: Systematic Review and Meta-Analysis.JMIR Med Inform. 2020 Nov 17;8(11):e16503. doi: 10.2196/16503. JMIR Med Inform. 2020. PMID: 33200995 Free PMC article. Review.
References
-
- Bar-Ami S, Gitay-Goren H. Increased progesterone secretion and 3 beta-hydroxysteroid dehydrogenase activity in human cumulus cells by pregnenolone is limited to the high steroidogenic active cumuli. Journal of assisted reproduction and genetics. 2000;17: 437–44. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3455565&tool=p... - PMC - PubMed
-
- Adriaenssens T, Wathlet S, Segers I, Verheyen G, De Vos A, Van der Elst J, et al. Cumulus cell gene expression is associated with oocyte developmental quality and influenced by patient and treatment characteristics. Human reproduction (Oxford, England). 2010;25: 1259–1270. 10.1093/humrep/deq049 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

