Reverse engineering GTPase programming languages with reconstituted signaling networks
- PMID: 27128855
- PMCID: PMC5003541
- DOI: 10.1080/21541248.2016.1178367
Reverse engineering GTPase programming languages with reconstituted signaling networks
Abstract
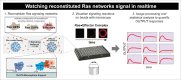
The Ras superfamily GTPases represent one of the most prolific signaling currencies used in Eukaryotes. With these remarkable molecules, evolution has built GTPase networks that control diverse cellular processes such as growth, morphology, motility and trafficking. (1-4) Our knowledge of the individual players that underlie the function of these networks is deep; decades of biochemical and structural data has provided a mechanistic understanding of the molecules that turn GTPases ON and OFF, as well as how those GTPase states signal by controlling the assembly of downstream effectors. However, we know less about how these different activities work together as a system to specify complex dynamic signaling outcomes. Decoding this molecular "programming language" would help us understand how different species and cell types have used the same GTPase machinery in different ways to accomplish different tasks, and would also provide new insights as to how mutations to these networks can cause disease. We recently developed a bead-based microscopy assay to watch reconstituted H-Ras signaling systems at work under arbitrary configurations of regulators and effectors. (5) Here we highlight key observations and insights from this study and propose extensions to our method to further study this and other GTPase signaling systems.
Keywords: Ras; biochemistry; dynamics; in vitro reconstitution; signaling.
Figures


References
-
- Chang F, Steelman LS, Shelton JG, Lee JT, Navolanic PM, Blalock WL, Franklin R, McCubrey JA. Regulation of cell cycle progression and apoptosis by the Ras/Raf/MEK/ERK pathway (Review). Int J Oncol 2003; 22:469-80; PMID:12579299. - PubMed
-
- Sjölander A, Yamamoto K, Huber BE, Lapetina EG. Association of p21ras with phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A 1991; 88:7908-12; PMID:1716764; http://dx.doi.org/10.1073/pnas.88.18.7908 - DOI - PMC - PubMed
-
- Hofer F, Fields S, Schneider C, Martin GS. Activated Ras interacts with the Ral guanine nucleotide dissociation stimulator. Proc Natl Acad Sci 1994; 91:11089-93; PMID:7972015; http://dx.doi.org/10.1073/pnas.91.23.11089 - DOI - PMC - PubMed
-
- Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature 1990; 348:125-32; PMID:2122258; http://dx.doi.org/10.1038/348125a0 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
