Neoadjuvant tamoxifen synchronizes ERα binding and gene expression profiles related to outcome and proliferation
- PMID: 27129152
- PMCID: PMC5085127
- DOI: 10.18632/oncotarget.8983
Neoadjuvant tamoxifen synchronizes ERα binding and gene expression profiles related to outcome and proliferation
Abstract
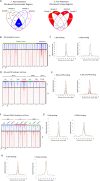
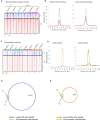
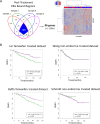
Estrogen receptor alpha (ERα)-positive breast cancers are frequently treated with tamoxifen, but resistance is common. It remains elusive how tamoxifen resistance occurs and predictive biomarkers for treatment outcome are needed. Because most biomarker discovery studies are performed using pre-treatment surgical resections, the effects of tamoxifen therapy directly on the tumor cell in vivo remain unexamined. In this study, we assessed DNA copy number, gene expression profiles and ERα/chromatin binding landscapes on breast tumor specimens, both before and after neoadjuvant tamoxifen treatment. We observed neoadjuvant tamoxifen treatment synchronized ERα/chromatin interactions and downstream gene expression, indicating that hormonal therapy reduces inter-tumor molecular variability. ERα-synchronized sites are associated with dynamic FOXA1 action at these sites, which is under control of growth factor signaling. Genes associated with tamoxifen-synchronized sites are capable of differentiating patients for tamoxifen benefit. Due to the direct effects of therapeutics on ERα behavior and transcriptional output, our study highlights the added value of biomarker discovery studies after neoadjuvant drug exposure.
Keywords: ChIP-seq; endocrine therapy; estrogen receptor; gene expression analysis; neoadjuvant therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer J Int Cancer. 2015 Mar 1;136:E359–86. - PubMed
-
- Fox MS. On the diagnosis and treatment of breast cancer. JAMA J Am Med Assoc. 1979 Feb 2;241:489–94. - PubMed
-
- Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, Wilfond B, Borg A, Trent J, et al. Gene-expression profiles in hereditary breast cancer. N Engl J Med. 2001 Feb 22;344:539–48. - PubMed
-
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, et al. Molecular portraits of human breast tumours. Nature. 2000 Aug 17;406:747–52. - PubMed
-
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001 Sep 11;98:10869–74. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

