Gut microbiome and kidney disease: a bidirectional relationship
- PMID: 27129691
- PMCID: PMC5399049
- DOI: 10.1007/s00467-016-3392-7
Gut microbiome and kidney disease: a bidirectional relationship
Abstract
Recent technological advances and efforts, including powerful metagenomic and metatranscriptomic analyses, have led to a tremendous growth in our understanding of microbial communities. Changes in microbial abundance or composition of human microbial communities impact human health or disease state. However, explorations into the mechanisms underlying host-microbe interactions in health and disease are still in their infancy. Although changes in the gut microbiota have been described in patients with kidney disease, the relationships between pathogenesis, mechanisms of disease progression, and the gut microbiome are still evolving. Here, we review changes in the host-microbiome symbiotic relationship in an attempt to explore the bidirectional relationship in which alterations in the microbiome affect kidney disease progression and how kidney disease may disrupt a balanced microbiome. We also discuss potential targeted interventions that may help re-establish this symbiosis and propose more effective ways to deploy traditional treatments in patients with kidney disease.
Keywords: Acute kidney injury; Chronic kidney disease; Dysbiosis; End-stage renal disease; Hemodialysis; Hypertension; Microbiota; Peritoneal dialysis; Prebiotics; Probiotics; Transplantation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

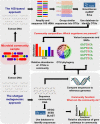
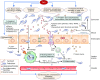
References
-
- Solt I, Kim MJ, Offer C. The human microbiome. Harefuah. 2011;150:484–488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

