Multi-omic data integration and analysis using systems genomics approaches: methods and applications in animal production, health and welfare
- PMID: 27130220
- PMCID: PMC4850674
- DOI: 10.1186/s12711-016-0217-x
Multi-omic data integration and analysis using systems genomics approaches: methods and applications in animal production, health and welfare
Abstract
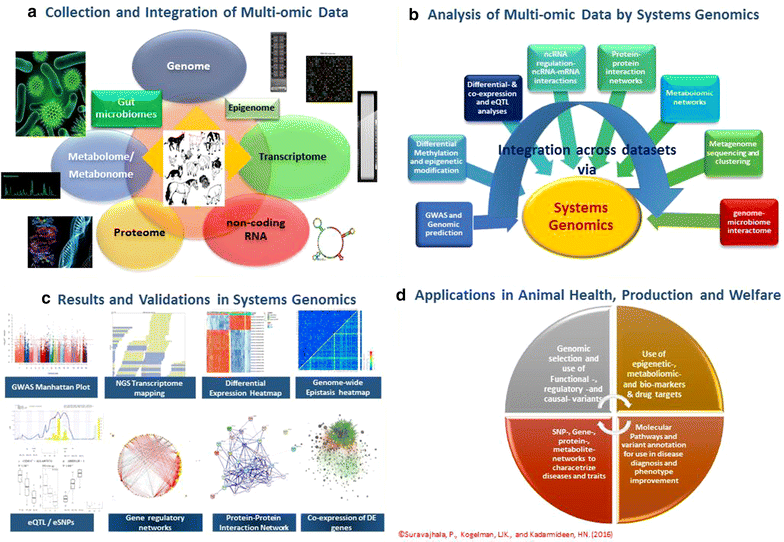
In the past years, there has been a remarkable development of high-throughput omics (HTO) technologies such as genomics, epigenomics, transcriptomics, proteomics and metabolomics across all facets of biology. This has spearheaded the progress of the systems biology era, including applications on animal production and health traits. However, notwithstanding these new HTO technologies, there remains an emerging challenge in data analysis. On the one hand, different HTO technologies judged on their own merit are appropriate for the identification of disease-causing genes, biomarkers for prevention and drug targets for the treatment of diseases and for individualized genomic predictions of performance or disease risks. On the other hand, integration of multi-omic data and joint modelling and analyses are very powerful and accurate to understand the systems biology of healthy and sustainable production of animals. We present an overview of current and emerging HTO technologies each with a focus on their applications in animal and veterinary sciences before introducing an integrative systems genomics framework for analysing and integrating multi-omic data towards improved animal production, health and welfare. We conclude that there are big challenges in multi-omic data integration, modelling and systems-level analyses, particularly with the fast emerging HTO technologies. We highlight existing and emerging systems genomics approaches and discuss how they contribute to our understanding of the biology of complex traits or diseases and holistic improvement of production performance, disease resistance and welfare.
Figures
References
-
- Kadarmideen HN. Genomics to systems biology in animal and veterinary sciences: progress, lessons and opportunities. Livest Sci. 2014;166:232–248. doi: 10.1016/j.livsci.2014.04.028. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

