miEAA: microRNA enrichment analysis and annotation
- PMID: 27131362
- PMCID: PMC4987907
- DOI: 10.1093/nar/gkw345
miEAA: microRNA enrichment analysis and annotation
Abstract
Similar to the development of gene set enrichment and gene regulatory network analysis tools over a decade ago, microRNA enrichment tools are currently gaining importance. Building on our experience with the gene set analysis toolkit GeneTrail, we implemented the miRNA Enrichment Analysis and Annotation tool (miEAA). MiEAA is a web-based application that offers a variety of commonly applied statistical tests such as over-representation analysis and miRNA set enrichment analysis, which is similar to Gene Set Enrichment Analysis. Besides the different statistical tests, miEAA also provides rich functionality in terms of miRNA categories. Altogether, over 14 000 miRNA sets have been added, including pathways, diseases, organs and target genes. Importantly, our tool can be applied for miRNA precursors as well as mature miRNAs. To make the tool as useful as possible we additionally implemented supporting tools such as converters between different miRBase versions and converters from miRNA names to precursor names. We evaluated the performance of miEAA on two sets of miRNAs that are affected in lung adenocarcinomas and have been detected by array analysis. The web-based application is freely accessible at: http://www.ccb.uni-saarland.de/mieaa_tool/.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
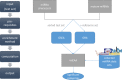
References
-
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. - PubMed
-
- Friedlander M.R., Chen W., Adamidi C., Maaskola J., Einspanier R., Knespel S., Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotech. 2008;26:407–415. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

