Mapping the Human Platelet Lipidome Reveals Cytosolic Phospholipase A2 as a Regulator of Mitochondrial Bioenergetics during Activation
- PMID: 27133131
- PMCID: PMC4873619
- DOI: 10.1016/j.cmet.2016.04.001
Mapping the Human Platelet Lipidome Reveals Cytosolic Phospholipase A2 as a Regulator of Mitochondrial Bioenergetics during Activation
Abstract
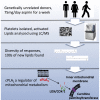
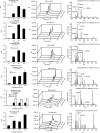
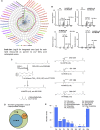
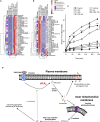
Human platelets acutely increase mitochondrial energy generation following stimulation. Herein, a lipidomic circuit was uncovered whereby the substrates for this are exclusively provided by cPLA2, including multiple fatty acids and oxidized species that support energy generation via β-oxidation. This indicates that acute lipid membrane remodeling is required to support energetic demands during platelet activation. Phospholipase activity is linked to energy metabolism, revealing cPLA2 as a central regulator of both lipidomics and energy flux. Using a lipidomic approach (LipidArrays), we also estimated the total number of lipids in resting, thrombin-activated, and aspirinized platelets. Significant diversity between genetically unrelated individuals and a wealth of species was revealed. Resting platelets demonstrated ∼5,600 unique species, with only ∼50% being putatively identified. Thrombin elevated ∼900 lipids >2-fold with 86% newly appearing and 45% inhibited by aspirin supplementation, indicating COX-1 is required for major activation-dependent lipidomic fluxes. Many lipids were structurally identified. With ∼50% of the lipids being absent from databases, a major opportunity for mining lipids relevant to human health and disease is presented.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



Comment in
-
Human Platelet Lipidomics: Variance, Visualization, Flux, and Fuel.Cell Metab. 2016 May 10;23(5):757-9. doi: 10.1016/j.cmet.2016.04.025. Cell Metab. 2016. PMID: 27166936
References
-
- Aldrovandi M., Hammond V.J., Podmore H., Hornshaw M., Clark S.R., Marnett L.J., Slatter D.A., Murphy R.C., Collins P.W., O’Donnell V.B. Human platelets generate phospholipid-esterified prostaglandins via cyclooxygenase-1 that are inhibited by low dose aspirin supplementation. J. Lipid Res. 2013;54:3085–3097. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

