Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt)
- PMID: 27133166
- PMCID: PMC4889216
- DOI: 10.1016/j.cell.2016.04.011
Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt)
Abstract
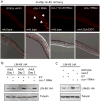
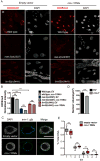
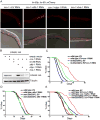
Organisms respond to mitochondrial stress through the upregulation of an array of protective genes, often perpetuating an early response to metabolic dysfunction across a lifetime. We find that mitochondrial stress causes widespread changes in chromatin structure through histone H3K9 di-methylation marks traditionally associated with gene silencing. Mitochondrial stress response activation requires the di-methylation of histone H3K9 through the activity of the histone methyltransferase met-2 and the nuclear co-factor lin-65. While globally the chromatin becomes silenced by these marks, remaining portions of the chromatin open up, at which point the binding of canonical stress responsive factors such as DVE-1 occurs. Thus, a metabolic stress response is established and propagated into adulthood of animals through specific epigenetic modifications that allow for selective gene expression and lifespan extension.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Mitochondria: Masters of Epigenetics.Cell. 2016 May 19;165(5):1052-1054. doi: 10.1016/j.cell.2016.05.021. Cell. 2016. PMID: 27203109 Free PMC article.
-
Ageing: The yin and yang of mitochondrial dysfunction.Nat Rev Mol Cell Biol. 2016 May 23;17(6):331. doi: 10.1038/nrm.2016.71. Nat Rev Mol Cell Biol. 2016. PMID: 27211483 No abstract available.
References
-
- Andersen EC, Horvitz HR. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Dev Camb Engl. 2007;134:2991–2999. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

