Structural Insights into Ring Formation of Cohesin and Related Smc Complexes
- PMID: 27134029
- PMCID: PMC4989898
- DOI: 10.1016/j.tcb.2016.04.002
Structural Insights into Ring Formation of Cohesin and Related Smc Complexes
Abstract
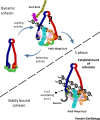
Cohesin facilitates sister chromatid cohesion through the formation of a large ring structure that encircles DNA. Its function relies on two structural maintenance of chromosomes (Smc) proteins, which are found in almost all organisms tested, from bacteria to humans. In accordance with their ubiquity, Smc complexes, such as cohesin, condensin, Smc5-6, and the dosage compensation complex, affect almost all processes of DNA homeostasis. Although their precise molecular mechanism remains enigmatic, here we provide an overview of the architecture of eukaryotic Smc complexes with a particular focus on cohesin, which has seen the most progress recently. Given the evident conservation of many structural features between Smc complexes, it is expected that architecture and topology will have a significant role when deciphering their precise molecular mechanisms.
Copyright © 2016 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures


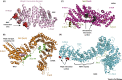

References
-
- Cobbe N., Heck M.M. The evolution of SMC proteins: phylogenetic analysis and structural implications. Mol. Biol. Evol. 2004;21:332–347. - PubMed
-
- Ames G.F., Lecar H. ATP-dependent bacterial transporters and cystic fibrosis: analogy between channels and transporters. FASEB J. 1992;6:2660–2666. - PubMed
-
- Cobbe N., Heck M.M. The evolution of ATPase activity in SMC proteins. Proteins. 2006;63:685–696. - PubMed
-
- Schleiffer A. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol. Cell. 2003;11:571–575. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

