Reverse transcription-PCR assays for the differentiation of various US porcine epidemic diarrhea virus strains
- PMID: 27134071
- PMCID: PMC7173223
- DOI: 10.1016/j.jviromet.2016.04.018
Reverse transcription-PCR assays for the differentiation of various US porcine epidemic diarrhea virus strains
Abstract
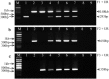
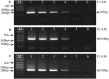
Concurrently, several porcine epidemic diarrhea virus (PEDV) variants are circulating in US swine farms, including the original US and the spike insertion-deletion (S-INDEL) strains. In this study, reverse transcription (RT)-PCR assays for the detection and differentiation of different US PEDV variants were developed based on the differences in the S1 domain of the spike (S) gene. This assay successfully differentiated three PEDV strains: PC22A (the original US virulent), Iowa106 (S-INDEL), and PC177 (S-197DEL) that was derived from cell culture adaptation and has a 197 amino acid-deletion in the S1 domain. The assays did not amplify the porcine deltacoronavirus OH-FD22 strain or transmissible gastroenteritis virus Miller strain. It is the first report on the development of RT-PCR assays allowing the detection and differentiation of all major types of US PEDV variants.
Keywords: Porcine epidemic diarrhea virus; Reverse transcription-PCR; Spike gene.
Copyright © 2016 Elsevier B.V. All rights reserved.
Figures


Similar articles
-
Deletion of a 197-Amino-Acid Region in the N-Terminal Domain of Spike Protein Attenuates Porcine Epidemic Diarrhea Virus in Piglets.J Virol. 2017 Jun 26;91(14):e00227-17. doi: 10.1128/JVI.00227-17. Print 2017 Jul 15. J Virol. 2017. PMID: 28490591 Free PMC article.
-
Detection and phylogenetic analyses of spike genes in porcine epidemic diarrhea virus strains circulating in China in 2016-2017.Virol J. 2017 Oct 10;14(1):194. doi: 10.1186/s12985-017-0860-z. Virol J. 2017. PMID: 29017599 Free PMC article.
-
Experimental infection of a US spike-insertion deletion porcine epidemic diarrhea virus in conventional nursing piglets and cross-protection to the original US PEDV infection.Vet Res. 2015 Nov 20;46:134. doi: 10.1186/s13567-015-0278-9. Vet Res. 2015. PMID: 26589292 Free PMC article.
-
Porcine epidemic diarrhea virus: An overview of current virological and serological diagnostic methods.Virus Res. 2016 Dec 2;226:60-70. doi: 10.1016/j.virusres.2016.05.013. Epub 2016 May 14. Virus Res. 2016. PMID: 27189041 Free PMC article. Review.
-
Porcine epidemic diarrhea: A retrospect from Europe and matters of debate.Virus Res. 2016 Dec 2;226:1-6. doi: 10.1016/j.virusres.2016.05.030. Epub 2016 Jun 15. Virus Res. 2016. PMID: 27317168 Free PMC article. Review.
Cited by
-
Rapid differentiation of PEDV wild-type strains and classical attenuated vaccine strains by fluorescent probe-based reverse transcription recombinase polymerase amplification assay.BMC Vet Res. 2020 Jun 22;16(1):208. doi: 10.1186/s12917-020-02424-1. BMC Vet Res. 2020. PMID: 32571305 Free PMC article.
-
Characterisation of porcine epidemic diarrhea virus isolates during the 2014-2015 outbreak in the Philippines.Virusdisease. 2018 Sep;29(3):342-348. doi: 10.1007/s13337-018-0470-4. Epub 2018 Jun 28. Virusdisease. 2018. PMID: 30159369 Free PMC article.
-
Identification of porcine epidemic diarrhea virus variant with a large spike gene deletion from a clinical swine sample in the United States.Virus Genes. 2018 Apr;54(2):323-327. doi: 10.1007/s11262-018-1542-7. Epub 2018 Feb 21. Virus Genes. 2018. PMID: 29468451 Free PMC article.
-
New variants of porcine epidemic diarrhea virus with large deletions in the spike protein, identified in the United States, 2016-2017.Arch Virol. 2018 Sep;163(9):2485-2489. doi: 10.1007/s00705-018-3874-y. Epub 2018 May 22. Arch Virol. 2018. PMID: 29789941 Free PMC article.
-
Advances in porcine epidemic diarrhea virus research: genome, epidemiology, vaccines, and detection methods.Discov Nano. 2025 Mar 3;20(1):48. doi: 10.1186/s11671-025-04220-y. Discov Nano. 2025. PMID: 40029472 Free PMC article. Review.
References
-
- Chen Q., Li G., Stasko J., Thomas J.T., Stensland W.R., Pillatzki A.E., Gauger P.C., Schwartz K.J., Madson D., Yoon K.J., Stevenson G.W., Burrough E.R., Harmon K.M., Main R.G., Zhang J. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014;52:234–243. doi: 10.1128/JCM.02820-13. - DOI - PMC - PubMed
-
- Debouck P., Pensaert M., Coussement W. The pathogenesis of an enteric infection in pigs, experimentally induced by the coronavirus-like agent, CV777. Vet. Microbiol. 1981;6:157–165.
-
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999:95–98. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

