Origin, fate and dynamics of macrophages at central nervous system interfaces
- PMID: 27135602
- PMCID: PMC4968048
- DOI: 10.1038/ni.3423
Origin, fate and dynamics of macrophages at central nervous system interfaces
Abstract
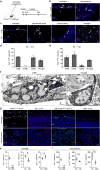
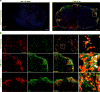
Perivascular, subdural meningeal and choroid plexus macrophages are non-parenchymal macrophages that mediate immune responses at brain boundaries. Although the origin of parenchymal microglia has recently been elucidated, much less is known about the precursors, the underlying transcriptional program and the dynamics of the other macrophages in the central nervous system (CNS). It was assumed that they have a high turnover from blood-borne monocytes. However, using parabiosis and fate-mapping approaches in mice, we found that CNS macrophages arose from hematopoietic precursors during embryonic development and established stable populations, with the notable exception of choroid plexus macrophages, which had dual origins and a shorter life span. The generation of CNS macrophages relied on the transcription factor PU.1, whereas the MYB, BATF3 and NR4A1 transcription factors were not required.
Figures





Comment in
-
Family ties among CNS macrophages.Nat Immunol. 2016 Jun 21;17(7):742-3. doi: 10.1038/ni.3490. Nat Immunol. 2016. PMID: 27327994 No abstract available.
References
-
- Ransohoff RM, Cardona AE. The myeloid cells of the central nervous system parenchyma. Nature. 2010;468:253–262. - PubMed
-
- Prinz M, Priller J, Sisodia SS, Ransohoff RM. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat Neurosci. 2011;14:1227–1235. - PubMed
-
- Prinz M, Priller J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci. 2014;15:300–312. - PubMed
-
- Alliot F, Godin I, Pessac B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res Dev Bra in Res. 1999;117:145–152. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

