Landscape of somatic mutations in 560 breast cancer whole-genome sequences
- PMID: 27135926
- PMCID: PMC4910866
- DOI: 10.1038/nature17676
Landscape of somatic mutations in 560 breast cancer whole-genome sequences
Erratum in
-
Author Correction: Landscape of somatic mutations in 560 breast cancer whole-genome sequences.Nature. 2019 Feb;566(7742):E1. doi: 10.1038/s41586-019-0883-2. Nature. 2019. PMID: 30659290
Abstract
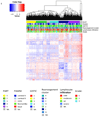
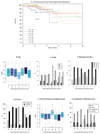
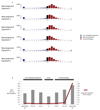
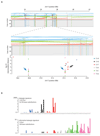
We analysed whole-genome sequences of 560 breast cancers to advance understanding of the driver mutations conferring clonal advantage and the mutational processes generating somatic mutations. We found that 93 protein-coding cancer genes carried probable driver mutations. Some non-coding regions exhibited high mutation frequencies, but most have distinctive structural features probably causing elevated mutation rates and do not contain driver mutations. Mutational signature analysis was extended to genome rearrangements and revealed twelve base substitution and six rearrangement signatures. Three rearrangement signatures, characterized by tandem duplications or deletions, appear associated with defective homologous-recombination-based DNA repair: one with deficient BRCA1 function, another with deficient BRCA1 or BRCA2 function, the cause of the third is unknown. This analysis of all classes of somatic mutation across exons, introns and intergenic regions highlights the repertoire of cancer genes and mutational processes operating, and progresses towards a comprehensive account of the somatic genetic basis of breast cancer.
Figures







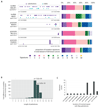
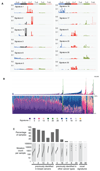
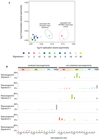

Comment in
-
Cancer genomics: A catalogue of somatic mutations.Nat Rev Genet. 2016 Jul;17(7):378. doi: 10.1038/nrg.2016.65. Epub 2016 May 9. Nat Rev Genet. 2016. PMID: 27156977 No abstract available.
-
Genetics: Defining driver mutations in the genomic landscape of breast cancer.Nat Rev Clin Oncol. 2016 May 20;13(6):327. doi: 10.1038/nrclinonc.2016.75. Nat Rev Clin Oncol. 2016. PMID: 27199176 No abstract available.
-
Elementary: breast cancer culprits leave their signatures on the double helix.Cell Death Differ. 2016 Oct;23(10):1577-8. doi: 10.1038/cdd.2016.82. Epub 2016 Sep 2. Cell Death Differ. 2016. PMID: 27588706 Free PMC article. No abstract available.
-
An ICGC major achievement in breast cancer: a comprehensive catalog of mutations and mutational signatures.Chin Clin Oncol. 2017 Feb;6(1):4. doi: 10.21037/cco.2016.11.01. Epub 2016 Nov 9. Chin Clin Oncol. 2017. PMID: 28061528 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- 322737/ERC_/European Research Council/International
- P50 CA168504/CA/NCI NIH HHS/United States
- WT100183MA/WT_/Wellcome Trust/United Kingdom
- 098051/WT_/Wellcome Trust/United Kingdom
- 100183/WT_/Wellcome Trust/United Kingdom
- 20952/CRUK_/Cancer Research UK/United Kingdom
- 077012/Z/05/Z/WT_/Wellcome Trust/United Kingdom
- 088177/WT_/Wellcome Trust/United Kingdom
- P30 CA008748/CA/NCI NIH HHS/United States
- P50 CA168504-02/CA/NCI NIH HHS/United States
- CSO_/Chief Scientist Office/United Kingdom
- 16942/CRUK_/Cancer Research UK/United Kingdom
- 077012/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

