Structural Insights into Protein-Protein Interactions Involved in Bacterial Cell Wall Biogenesis
- PMID: 27136593
- PMCID: PMC4929429
- DOI: 10.3390/antibiotics5020014
Structural Insights into Protein-Protein Interactions Involved in Bacterial Cell Wall Biogenesis
Abstract
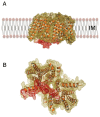
The bacterial cell wall is essential for survival, and proteins that participate in its biosynthesis have been the targets of antibiotic development efforts for decades. The biosynthesis of its main component, the peptidoglycan, involves the coordinated action of proteins that are involved in multi-member complexes which are essential for cell division (the "divisome") and/or cell wall elongation (the "elongasome"), in the case of rod-shaped cells. Our knowledge regarding these interactions has greatly benefitted from the visualization of different aspects of the bacterial cell wall and its cytoskeleton by cryoelectron microscopy and tomography, as well as genetic and biochemical screens that have complemented information from high resolution crystal structures of protein complexes involved in divisome or elongasome formation. This review summarizes structural and functional aspects of protein complexes involved in the cytoplasmic and membrane-related steps of peptidoglycan biosynthesis, with a particular focus on protein-protein interactions whereby disruption could lead to the development of novel antibacterial strategies.
Keywords: MraY; Mur enzymes; bacterial cytoskeleton; cell division; elongation; peptidoglycan; protein complexes.
Figures


Similar articles
-
Do the divisome and elongasome share a common evolutionary past?Curr Opin Microbiol. 2013 Dec;16(6):745-51. doi: 10.1016/j.mib.2013.09.003. Epub 2013 Oct 1. Curr Opin Microbiol. 2013. PMID: 24094808 Review.
-
Penicillin-Binding Proteins (PBPs) and Bacterial Cell Wall Elongation Complexes.Subcell Biochem. 2019;93:273-289. doi: 10.1007/978-3-030-28151-9_8. Subcell Biochem. 2019. PMID: 31939154 Review.
-
Molecular architecture of the PBP2-MreC core bacterial cell wall synthesis complex.Nat Commun. 2017 Oct 3;8(1):776. doi: 10.1038/s41467-017-00783-2. Nat Commun. 2017. PMID: 28974686 Free PMC article.
-
Bacterial cell division proteins as antibiotic targets.Bioorg Chem. 2014 Aug;55:27-38. doi: 10.1016/j.bioorg.2014.03.007. Epub 2014 Apr 4. Bioorg Chem. 2014. PMID: 24755375 Review.
-
Class A PBPs have a distinct and unique role in the construction of the pneumococcal cell wall.Proc Natl Acad Sci U S A. 2020 Mar 17;117(11):6129-6138. doi: 10.1073/pnas.1917820117. Epub 2020 Mar 2. Proc Natl Acad Sci U S A. 2020. PMID: 32123104 Free PMC article.
Cited by
-
Analysis of variations in cell envelope subproteome and cell length in Acinetobacter baumannii ATCC 19606T populations by effect of temperature and desiccation.Int Microbiol. 2025 Aug 23. doi: 10.1007/s10123-025-00706-y. Online ahead of print. Int Microbiol. 2025. PMID: 40846826
-
In Cellulo Synthesis of Proteins Containing a Fluorescent Oxazole Amino Acid.J Am Chem Soc. 2019 Apr 10;141(14):5597-5601. doi: 10.1021/jacs.8b12767. Epub 2019 Mar 26. J Am Chem Soc. 2019. PMID: 30889951 Free PMC article.
-
Determinants of Bacterial Morphology: From Fundamentals to Possibilities for Antimicrobial Targeting.Front Microbiol. 2017 Jul 10;8:1264. doi: 10.3389/fmicb.2017.01264. eCollection 2017. Front Microbiol. 2017. PMID: 28740487 Free PMC article. Review.
-
Arginine methylation sites on SepIVA help balance elongation and septation in Mycobacterium smegmatis.Mol Microbiol. 2023 Feb;119(2):208-223. doi: 10.1111/mmi.15006. Epub 2022 Dec 4. Mol Microbiol. 2023. PMID: 36416406 Free PMC article.
-
Zingiber officinale and Vernonia amygdalina Infusions Improve Redox Status in Rat Brain.Evid Based Complement Alternat Med. 2022 Sep 26;2022:9470178. doi: 10.1155/2022/9470178. eCollection 2022. Evid Based Complement Alternat Med. 2022. PMID: 36199544 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

