Identification of Chimeric Repressors that Confer Salt and Osmotic Stress Tolerance in Arabidopsis
- PMID: 27137403
- PMCID: PMC4844390
- DOI: 10.3390/plants2040769
Identification of Chimeric Repressors that Confer Salt and Osmotic Stress Tolerance in Arabidopsis
Abstract
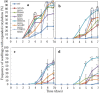
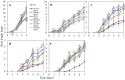
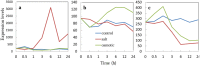
We produced transgenic Arabidopsis plants that express chimeric genes for transcription factors converted to dominant repressors, using Chimeric REpressor gene-Silencing Technology (CRES-T), and evaluated the salt tolerance of each line. The seeds of the CRES-T lines for ADA2b, Msantd, DDF1, DREB26, AtGeBP, and ATHB23 exhibited higher germination rates than Wild type (WT) and developed rosette plants under up to 200 mM NaCl or 400 mM mannitol. WT plants did not grow under these conditions. In these CRES-T lines, the expression patterns of stress-related genes such as RD29A, RD22, DREB1A, and P5CS differed from those in WT plants, suggesting the involvement of the six transcription factors identified here in the stress response pathways regulated by the products of these stress-related genes. Our results demonstrate additional proof that CRES-T is a superior tool for revealing the function of transcription factors.
Keywords: CRES-T; osmotic stress; repressor; salt stress; stress tolerance; transcription factors.
Figures








References
-
- Shi H., Lee B.H., Wu S.J., Zhu J.K. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2003;21:81–85. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

