Ezrin Inhibition Up-regulates Stress Response Gene Expression
- PMID: 27137931
- PMCID: PMC4933238
- DOI: 10.1074/jbc.M116.718189
Ezrin Inhibition Up-regulates Stress Response Gene Expression
Abstract
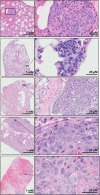
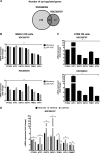
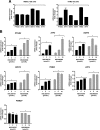
Ezrin is a member of the ERM (ezrin/radixin/moesin) family of proteins that links cortical cytoskeleton to the plasma membrane. High expression of ezrin correlates with poor prognosis and metastasis in osteosarcoma. In this study, to uncover specific cellular responses evoked by ezrin inhibition that can be used as a specific pharmacodynamic marker(s), we profiled global gene expression in osteosarcoma cells after treatment with small molecule ezrin inhibitors, NSC305787 and NSC668394. We identified and validated several up-regulated integrated stress response genes including PTGS2, ATF3, DDIT3, DDIT4, TRIB3, and ATF4 as novel ezrin-regulated transcripts. Analysis of transcriptional response in skin and peripheral blood mononuclear cells from NSC305787-treated mice compared with a control group revealed that, among those genes, the stress gene DDIT4/REDD1 may be used as a surrogate pharmacodynamic marker of ezrin inhibitor compound activity. In addition, we validated the anti-metastatic effects of NSC305787 in reducing the incidence of lung metastasis in a genetically engineered mouse model of osteosarcoma and evaluated the pharmacokinetics of NSC305787 and NSC668394 in mice. In conclusion, our findings suggest that cytoplasmic ezrin, previously considered a dormant and inactive protein, has important functions in regulating gene expression that may result in down-regulation of stress response genes.
Keywords: ezrin; gene expression; metastasis; osteosarcoma; small molecule; transgenic mice.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Bretscher A., Edwards K., and Fehon R. G. (2002) ERM proteins and merlin: integrators at the cell cortex. Nat. Rev. Mol. Cell Biol. 3, 586–599 - PubMed
-
- Federici C., Brambilla D., Lozupone F., Matarrese P., de Milito A., Lugini L., Iessi E., Cecchetti S., Marino M., Perdicchio M., Logozzi M., Spada M., Malorni W., and Fais S. (2009) Pleiotropic function of ezrin in human metastatic melanomas. Int. J. Cancer 124, 2804–2812 - PubMed
-
- Köbel M., Langhammer T., Hüttelmaier S., Schmitt W. D., Kriese K., Dittmer J., Strauss H. G., Thomssen C., and Hauptmann S. (2006) Ezrin expression is related to poor prognosis in FIGO stage I endometrioid carcinomas. Mod. Pathol. 19, 581–587 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

