The genetic basis for ecological adaptation of the Atlantic herring revealed by genome sequencing
- PMID: 27138043
- PMCID: PMC4854517
- DOI: 10.7554/eLife.12081
The genetic basis for ecological adaptation of the Atlantic herring revealed by genome sequencing
Abstract
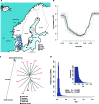
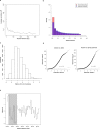
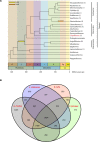
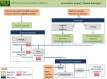
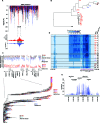
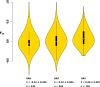
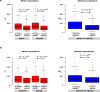
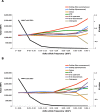
Ecological adaptation is of major relevance to speciation and sustainable population management, but the underlying genetic factors are typically hard to study in natural populations due to genetic differentiation caused by natural selection being confounded with genetic drift in subdivided populations. Here, we use whole genome population sequencing of Atlantic and Baltic herring to reveal the underlying genetic architecture at an unprecedented detailed resolution for both adaptation to a new niche environment and timing of reproduction. We identify almost 500 independent loci associated with a recent niche expansion from marine (Atlantic Ocean) to brackish waters (Baltic Sea), and more than 100 independent loci showing genetic differentiation between spring- and autumn-spawning populations irrespective of geographic origin. Our results show that both coding and non-coding changes contribute to adaptation. Haplotype blocks, often spanning multiple genes and maintained by selection, are associated with genetic differentiation.
Keywords: Atlantic herring; evolutionary biology; genetic adaptation; genomics; natural selection.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures




References
-
- Amemiya CT, Alföldi J, Lee AP, Fan S, Philippe H, Maccallum I, Braasch I, Manousaki T, Schneider I, Rohner N, Organ C, Chalopin D, Smith JJ, Robinson M, Dorrington RA, Gerdol M, Aken B, Biscotti MA, Barucca M, Baurain D, Berlin AM, Blatch GL, Buonocore F, Burmester T, Campbell MS, Canapa A, Cannon JP, Christoffels A, De Moro G, Edkins AL, Fan L, Fausto AM, Feiner N, Forconi M, Gamieldien J, Gnerre S, Gnirke A, Goldstone JV, Haerty W, Hahn ME, Hesse U, Hoffmann S, Johnson J, Karchner SI, Kuraku S, Lara M, Levin JZ, Litman GW, Mauceli E, Miyake T, Mueller MG, Nelson DR, Nitsche A, Olmo E, Ota T, Pallavicini A, Panji S, Picone B, Ponting CP, Prohaska SJ, Przybylski D, Saha NR, Ravi V, Ribeiro FJ, Sauka-Spengler T, Scapigliati G, Searle SM, Sharpe T, Simakov O, Stadler PF, Stegeman JJ, Sumiyama K, Tabbaa D, Tafer H, Turner-Maier J, van Heusden P, White S, Williams L, Yandell M, Brinkmann H, Volff JN, Tabin CJ, Shubin N, Schartl M, Jaffe DB, Postlethwait JH, Venkatesh B, Di Palma F, Lander ES, Meyer A, Lindblad-Toh K. The african coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496:311–316. doi: 10.1038/nature12027. - DOI - PMC - PubMed
-
- Andersson L, Ryman N, Rosenberg R, Ståhl G. Genetic variability in atlantic herring (Clupea harengus harengus): Description of protein loci and population data. Hereditas. 1981;95:69–78. doi: 10.1111/j.1601-5223.1981.tb01330.x. - DOI
-
- Andrén T, Björck S, Andrén E, Conley D, Zillén L, Anjar J. The Development of the Baltic Sea Basin During the Last 130 ka. In: Harff J, Bjorck S, Hoth P, editors. The Baltic Sea Basin. Berlin, Heidelberg: Springer Berlin Heidelberg; 2011. pp. 75–97. - DOI
-
- Aneer G. Some speculations about the Baltic herring (Clupea harengus membras) in connection with the eutrophication of the Baltic Sea. Canadian Journal of Fisheries and Aquatic Sciences. 1985;42:s83–s90. doi: 10.1139/f85-264. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

