Efficient and Controlled Generation of 2D and 3D Bile Duct Tissue from Human Pluripotent Stem Cell-Derived Spheroids
- PMID: 27138846
- PMCID: PMC6186008
- DOI: 10.1007/s12015-016-9657-5
Efficient and Controlled Generation of 2D and 3D Bile Duct Tissue from Human Pluripotent Stem Cell-Derived Spheroids
Abstract
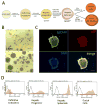
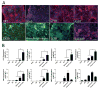
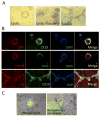
While in vitro liver tissue engineering has been increasingly studied during the last several years, presently engineered liver tissues lack the bile duct system. The lack of bile drainage not only hinders essential digestive functions of the liver, but also leads to accumulation of bile that is toxic to hepatocytes and known to cause liver cirrhosis. Clearly, generation of bile duct tissue is essential for engineering functional and healthy liver. Differentiation of human induced pluripotent stem cells (iPSCs) to bile duct tissue requires long and/or complex culture conditions, and has been inefficient so far. Towards generating a fully functional liver containing biliary system, we have developed defined and controlled conditions for efficient 2D and 3D bile duct epithelial tissue generation. A marker for multipotent liver progenitor in both adult human liver and ductal plate in human fetal liver, EpCAM, is highly expressed in hepatic spheroids generated from human iPSCs. The EpCAM high hepatic spheroids can, not only efficiently generate a monolayer of biliary epithelial cells (cholangiocytes), in a 2D differentiation condition, but also form functional ductal structures in a 3D condition. Importantly, this EpCAM high spheroid based biliary tissue generation is significantly faster than other existing methods and does not require cell sorting. In addition, we show that a knock-in CK7 reporter human iPSC line generated by CRISPR/Cas9 genome editing technology greatly facilitates the analysis of biliary differentiation. This new ductal differentiation method will provide a more efficient method of obtaining bile duct cells and tissues, which may facilitate engineering of complete and functional liver tissue in the future.
Keywords: 3D tissue engineering; Ductal differentiation; Genome editing; Induced pluripotent stem cells; Liver progenitor; Spheroids.
Conflict of interest statement
Conflict of Interest: The authors declare no potential conflicts of interest.
Figures

Similar articles
-
Functional Maturation of Induced Pluripotent Stem Cell Hepatocytes in Extracellular Matrix-A Comparative Analysis of Bioartificial Liver Microenvironments.Stem Cells Transl Med. 2016 Sep;5(9):1257-67. doi: 10.5966/sctm.2015-0235. Epub 2016 Jul 15. Stem Cells Transl Med. 2016. PMID: 27421950 Free PMC article.
-
Culture System of Bile Duct-Like Cystic Structures Derived from Human-Inducible Pluripotent Stem Cells.Methods Mol Biol. 2019;1905:143-153. doi: 10.1007/978-1-4939-8961-4_13. Methods Mol Biol. 2019. PMID: 30536097
-
Development and characterization of human-induced pluripotent stem cell-derived cholangiocytes.Lab Invest. 2015 Jun;95(6):684-96. doi: 10.1038/labinvest.2015.51. Epub 2015 Apr 13. Lab Invest. 2015. PMID: 25867762 Free PMC article.
-
Cholangiocytes derived from induced pluripotent stem cells for disease modeling.Curr Opin Gastroenterol. 2016 May;32(3):210-5. doi: 10.1097/MOG.0000000000000260. Curr Opin Gastroenterol. 2016. PMID: 27054776 Review.
-
Current strategies to generate mature human induced pluripotent stem cells derived cholangiocytes and future applications.Organogenesis. 2017 Jan 2;13(1):1-15. doi: 10.1080/15476278.2016.1278133. Epub 2017 Jan 5. Organogenesis. 2017. PMID: 28055309 Free PMC article. Review.
Cited by
-
3D Printed Model of Extrahepatic Biliary Ducts for Biliary Stent Testing.Materials (Basel). 2020 Oct 27;13(21):4788. doi: 10.3390/ma13214788. Materials (Basel). 2020. PMID: 33120964 Free PMC article.
-
Convergence of human pluripotent stem cell, organoid, and genome editing technologies.Exp Biol Med (Maywood). 2021 Apr;246(7):861-875. doi: 10.1177/1535370220985808. Epub 2021 Jan 19. Exp Biol Med (Maywood). 2021. PMID: 33467883 Free PMC article. Review.
-
Organoids and Spheroids as Models for Studying Cholestatic Liver Injury and Cholangiocarcinoma.Hepatology. 2021 Jul;74(1):491-502. doi: 10.1002/hep.31653. Epub 2021 Jun 4. Hepatology. 2021. PMID: 33222247 Free PMC article. Review.
-
Advanced Nanotheranostics of CRISPR/Cas for Viral Hepatitis and Hepatocellular Carcinoma.Adv Sci (Weinh). 2021 Dec;8(24):e2102051. doi: 10.1002/advs.202102051. Epub 2021 Oct 19. Adv Sci (Weinh). 2021. PMID: 34665528 Free PMC article. Review.
-
CRISPR/Cas9: at the cutting edge of hepatology.Gut. 2017 Jul;66(7):1329-1340. doi: 10.1136/gutjnl-2016-313565. Epub 2017 May 9. Gut. 2017. PMID: 28487442 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

