Single-cell transcriptional analysis of normal, aberrant, and malignant hematopoiesis in zebrafish
- PMID: 27139488
- PMCID: PMC4886368
- DOI: 10.1084/jem.20152013
Single-cell transcriptional analysis of normal, aberrant, and malignant hematopoiesis in zebrafish
Abstract
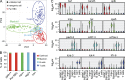
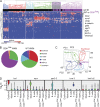
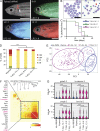
Hematopoiesis culminates in the production of functionally heterogeneous blood cell types. In zebrafish, the lack of cell surface antibodies has compelled researchers to use fluorescent transgenic reporter lines to label specific blood cell fractions. However, these approaches are limited by the availability of transgenic lines and fluorescent protein combinations that can be distinguished. Here, we have transcriptionally profiled single hematopoietic cells from zebrafish to define erythroid, myeloid, B, and T cell lineages. We also used our approach to identify hematopoietic stem and progenitor cells and a novel NK-lysin 4(+) cell type, representing a putative cytotoxic T/NK cell. Our platform also quantified hematopoietic defects in rag2(E450fs) mutant fish and showed that these fish have reduced T cells with a subsequent expansion of NK-lysin 4(+) cells and myeloid cells. These data suggest compensatory regulation of the innate immune system in rag2(E450fs) mutant zebrafish. Finally, analysis of Myc-induced T cell acute lymphoblastic leukemia showed that cells are arrested at the CD4(+)/CD8(+) cortical thymocyte stage and that a subset of leukemia cells inappropriately reexpress stem cell genes, including bmi1 and cmyb In total, our experiments provide new tools and biological insights into single-cell heterogeneity found in zebrafish blood and leukemia.
© 2016 Moore et al.
Figures



References
-
- Amir el A.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C., Shenfeld D.K., Krishnaswamy S., Nolan G.P., and Pe’er D.. 2013. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31:545–552. 10.1038/nbt.2594 - DOI - PMC - PubMed
-
- Andersson M., Gunne H., Agerberth B., Boman A., Bergman T., Sillard R., Jörnvall H., Mutt V., Olsson B., Wigzell H., et al. . 1995. NK-lysin, a novel effector peptide of cytotoxic T and NK cells. Structure and cDNA cloning of the porcine form, induction by interleukin 2, antibacterial and antitumour activity. EMBO J. 14:1615–1625. - PMC - PubMed
-
- Bendall S.C., Simonds E.F., Qiu P., Amir el A.D., Krutzik P.O., Finck R., Bruggner R.V., Melamed R., Trejo A., Ornatsky O.I., et al. . 2011. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 332:687–696. 10.1126/science.1198704 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

