Intragraft Antiviral-Specific Gene Expression as a Distinctive Transcriptional Signature for Studies in Polyomavirus-Associated Nephropathy
- PMID: 27140517
- PMCID: PMC5235336
- DOI: 10.1097/TP.0000000000001214
Intragraft Antiviral-Specific Gene Expression as a Distinctive Transcriptional Signature for Studies in Polyomavirus-Associated Nephropathy
Abstract
Background: Polyomavirus nephropathy (PVAN) is a common cause of kidney allograft dysfunction and loss. To identify PVAN-specific gene expression and underlying molecular mechanisms, we analyzed kidney biopsies with and without PVAN.
Methods: The study included 168 posttransplant renal allograft biopsies (T cell-mediated rejection [TCMR] = 26, PVAN = 10, normal functioning graft = 73, and interstitial fibrosis/tubular atrophy = 59) from 168 unique kidney allograft recipients. We performed gene expression assays and bioinformatics analysis to identify a set of PVAN-specific genes. Validity and relevance of a subset of these genes are validated by quantitative polymerase chain reaction and immunohistochemistry.
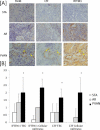
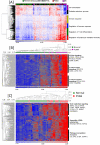
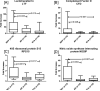
Results: Unsupervised hierarchical clustering analysis of all the biopsies revealed high similarity between PVAN and TCMR gene expression. Increased statistical stringency identified 158 and 252 unique PVAN and TCMR injury-specific gene transcripts respectively. Although TCMR-specific genes were overwhelmingly involved in immune response costimulation and TCR signaling, PVAN-specific genes were mainly related to DNA replication process, RNA polymerase assembly, and pathogen recognition receptors. A principal component analysis (PCA) using these genes further confirmed the most optimal separation between the 3 different clinical phenotypes. Validation of 4 PVAN-specific genes (RPS15, complement factor D, lactotransferrin, and nitric oxide synthase interacting protein) by quantitative polymerase chain reaction and confirmation by immunohistochemistry of 2 PVAN-specific proteins with antiviral function (lactotransferrin and IFN-inducible transmembrane 1) was done.
Conclusions: In conclusion, even though PVAN and TCMR kidney allografts share great similarities on gene perturbation, PVAN-specific genes were identified with well-known antiviral properties that provide tools for discerning PVAN and AR as well as attractive targets for rational drug design.
Figures

Similar articles
-
Predominant Tubular Interleukin-18 Expression in Polyomavirus-Associated Nephropathy.Transplantation. 2016 Oct;100(10):e88-95. doi: 10.1097/TP.0000000000001086. Transplantation. 2016. PMID: 26863474
-
Polyomavirus polymerase chain reaction as a surrogate marker of polyomavirus-associated nephropathy.Transplantation. 2007 Aug 15;84(3):340-5. doi: 10.1097/01.tp.0000275205.41078.51. Transplantation. 2007. PMID: 17700158
-
Clinical utility of histological features of polyomavirus allograft nephropathy.Transplantation. 2006 Jul 27;82(2):196-204. doi: 10.1097/01.tp.0000226176.87700.a4. Transplantation. 2006. PMID: 16858282
-
Polyomavirus-associated nephropathy: update in diagnosis.Transpl Infect Dis. 2006 Jun;8(2):68-75. doi: 10.1111/j.1399-3062.2006.00154.x. Transpl Infect Dis. 2006. PMID: 16734629 Review.
-
Polyomavirus-associated nephropathy in renal transplantation: interdisciplinary analyses and recommendations.Transplantation. 2005 May 27;79(10):1277-86. doi: 10.1097/01.tp.0000156165.83160.09. Transplantation. 2005. PMID: 15912088 Review.
Cited by
-
Fine needle aspirates of kidneys: a promising tool for RNA sequencing in native and transplanted kidneys.BMC Nephrol. 2018 Sep 5;19(1):221. doi: 10.1186/s12882-018-1012-4. BMC Nephrol. 2018. PMID: 30185151 Free PMC article.
-
Polyomavirus BK Nephropathy-Associated Transcriptomic Signatures: A Critical Reevaluation.Transplant Direct. 2018 Feb 2;4(2):e339. doi: 10.1097/TXD.0000000000000752. eCollection 2018 Feb. Transplant Direct. 2018. PMID: 29464200 Free PMC article.
-
Exploring the mechanism of BK polyomavirus-associated nephropathy through consensus gene network approach.PLoS One. 2023 Jun 15;18(6):e0282534. doi: 10.1371/journal.pone.0282534. eCollection 2023. PLoS One. 2023. PMID: 37319163 Free PMC article.
-
Defining housekeeping genes suitable for RNA-seq analysis of the human allograft kidney biopsy tissue.BMC Med Genomics. 2019 Jun 17;12(1):86. doi: 10.1186/s12920-019-0538-z. BMC Med Genomics. 2019. PMID: 31208411 Free PMC article.
-
BK Virus-Associated Nephropathy after Renal Transplantation.Pathogens. 2021 Feb 2;10(2):150. doi: 10.3390/pathogens10020150. Pathogens. 2021. PMID: 33540802 Free PMC article. Review.
References
-
- Dharnidharka VR, Cherikh WS, Abbott KC. An OPTN analysis of national registry data on treatment of BK virus allograft nephropathy in the United States. Transplantation. 2009;87(7):1019–1026. - PubMed
-
- Hirsch HH, Randhawa P. Practice ASTIDCo. BK polyomavirus in solid organ transplantation. Am J Tranplant. 2013;13(Suppl 4):179–188. - PubMed
-
- Ramos E, Drachenberg CB, Wali R, Hirsch HH. The decade of polyomavirus BK-associated nephropathy: state of affairs. Transplantation. 2009;87(5):621–630. - PubMed
-
- Hirsch HH, Knowles W, Dickenmann M, et al. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N J Engl Med. 2002;347(7):488–496. - PubMed
-
- Babel N, Volk HD, Reinke P. BK polyomavirus infection and nephropathy: the virus-immune system interplay. Nat Rev Nephrol. 2011;7(7):399–406. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

