Riches of phenotype computationally extracted from microbial colonies
- PMID: 27140647
- PMCID: PMC4878510
- DOI: 10.1073/pnas.1523295113
Riches of phenotype computationally extracted from microbial colonies
Abstract
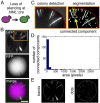
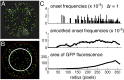
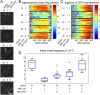
The genetic, epigenetic, and physiological differences among cells in clonal microbial colonies are underexplored opportunities for discovery. A recently developed genetic assay reveals that transient losses of heterochromatic repression, a heritable form of gene silencing, occur throughout the growth of Saccharomyces colonies. This assay requires analyzing two-color fluorescence patterns in yeast colonies, which is qualitatively appealing but quantitatively challenging. In this paper, we developed a suite of automated image processing, visualization, and classification algorithms (MORPHE) that facilitated the analysis of heterochromatin dynamics in the context of colonial growth and that can be broadly adapted to many colony-based assays in Saccharomyces and other microbes. Using the features that were automatically extracted from fluorescence images, our classification method distinguished loss-of-silencing patterns between mutants and wild type with unprecedented precision. Application of MORPHE revealed subtle but significant differences in the stability of heterochromatic repression between various environmental conditions, revealed that haploid cells experienced higher rates of silencing loss than diploids, and uncovered the unexpected contribution of a sirtuin to heterochromatin dynamics.
Keywords: epigenetics; feature extraction; heterochromatin dynamics; image segmentation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Meunier JR, Choder M. Saccharomyces cerevisiae colony growth and ageing: Biphasic growth accompanied by changes in gene expression. Yeast. 1999;15(12):1159–1169. - PubMed
-
- Váchová L, Kucerová H, Devaux F, Ulehlová M, Palková Z. Metabolic diversification of cells during the development of yeast colonies. Environ Microbiol. 2009;11(2):494–504. - PubMed
-
- Shapiro JA. The use of Mudlac transposons as tools for vital staining to visualize clonal and non-clonal patterns of organization in bacterial growth on agar surfaces. J Gen Microbiol. 1984;130(5):1169–1181. - PubMed
-
- Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56(5):771–776. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

