Maintenance of Epigenetic Information
- PMID: 27141050
- PMCID: PMC4852805
- DOI: 10.1101/cshperspect.a019372
Maintenance of Epigenetic Information
Abstract
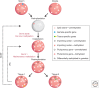
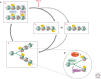
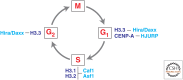
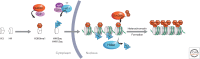
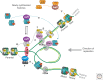
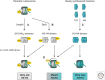
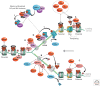
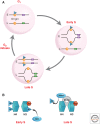
The genome is subject to a diverse array of epigenetic modifications from DNA methylation to histone posttranslational changes. Many of these marks are somatically stable through cell division. This article focuses on our knowledge of the mechanisms governing the inheritance of epigenetic marks, particularly, repressive ones, when the DNA and chromatin template are duplicated in S phase. This involves the action of histone chaperones, nucleosome-remodeling enzymes, histone and DNA methylation binding proteins, and chromatin-modifying enzymes. Last, the timing of DNA replication is discussed, including the question of whether this constitutes an epigenetic mark that facilitates the propagation of epigenetic marks.
Copyright © 2016 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures
References
-
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, et al. 1999. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J 18: 1923–1938. - PMC - PubMed
-
- Agalioti T, Chen G, Thanos D. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111: 381–392. - PubMed
-
- Ahmad K, Henikoff S. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9: 1191–1200. - PubMed
-
- Aladjem MI, Rodewald LW, Kolman JL, Wahl GM. 1998. Genetic dissection of a mammalian replicator in the human β-globin locus. Science 281: 1005–1009. - PubMed
-
- Allis CD, Jenuwein T, Reinberg D. 2014. Overview and concepts. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018739. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
