Epithelial ovarian cancer: the molecular genetics of epithelial ovarian cancer
- PMID: 27141069
- PMCID: PMC4852274
- DOI: 10.1093/annonc/mdw083
Epithelial ovarian cancer: the molecular genetics of epithelial ovarian cancer
Abstract
Background: Epithelial ovarian cancer (EOC) remains one of the leading causes of cancer-related deaths among women worldwide, despite gains in diagnostics and treatments made over the last three decades. Existing markers of ovarian cancer possess very limited clinical relevance highlighting the emerging need for identification of novel prognostic biomarkers as well as better predictive factors that might allow the stratification of patients who could benefit from a more targeted approach.
Patients and methods: A summary of molecular genetics of EOC.
Results: Large-scale high-throughput genomic technologies appear to be powerful tools for investigations into the genetic abnormalities in ovarian tumors, including studies on dysregulated genes and aberrantly activated signaling pathways. Such technologies can complement well-established clinical histopathology analysis and tumor grading and will hope to result in better, more tailored treatments in the future. Genomic signatures obtained by gene expression profiling of EOC may be able to predict survival outcomes and other important clinical outcomes, such as the success of surgical treatment. Finally, genomic analyses may allow for the identification of novel predictive biomarkers for purposes of treatment planning. These data combined suggest a pathway to progress in the treatment of advanced ovarian cancer and the promise of fulfilling the objective of providing personalized medicine to women with ovarian cancer.
Conclusions: The understanding of basic molecular events in the tumorigenesis and chemoresistance of EOC together with discovery of potential biomarkers may be greatly enhanced through large-scale genomic studies. In order to maximize the impact of these technologies, however, extensive validation studies are required.
Keywords: biomarkers; clinical trials; genomics; ovarian cancer.
© The Author 2016. Published by Oxford University Press on behalf of the European Society for Medical Oncology. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures
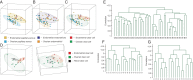

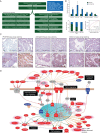

References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015; 65(1): 5–29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

