A Contra Capture Protein Array Platform for Studying Post-translationally Modified (PTM) Auto-antigenomes
- PMID: 27141097
- PMCID: PMC4937507
- DOI: 10.1074/mcp.M115.057661
A Contra Capture Protein Array Platform for Studying Post-translationally Modified (PTM) Auto-antigenomes
Abstract
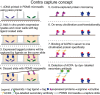
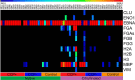
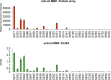
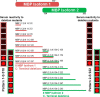
Aberrant modifications of proteins occur during disease development and elicit disease-specific antibody responses. We have developed a protein array platform that enables the modification of many proteins in parallel and assesses their immunogenicity without the need to express, purify, and modify proteins individually. We used anticitrullinated protein antibodies (ACPAs) in rheumatoid arthritis (RA) as a model modification and profiled antibody responses to ∼190 citrullinated proteins in 20 RA patients. We observed unique antibody reactivity patterns in both clinical anticyclic citrullinated peptide assay positive (CCP+) and CCP- RA patients. At individual antigen levels, we detected antibodies against known citrullinated autoantigens and discovered and validated five novel antibodies against specific citrullinated antigens (osteopontin (SPP1), flap endonuclease (FEN1), insulin like growth factor binding protein 6 (IGFBP6), insulin like growth factor I (IGF1) and stanniocalcin-2 (STC2)) in RA patients. We also demonstrated the utility of our innovative array platform in the identification of immune-dominant epitope(s) for citrullinated antigens. We believe our platform will promote the study of post-translationally modified antigens at a breadth that has not been achieved before, by both identifying novel autoantigens and investigating their roles in disease development. The developed platforms can potentially be used to study many autoimmune disease-relevant modifications and their immunogenicity.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
Monoclonal antibody against citrullinated peptides obtained from rheumatoid arthritis patients reacts with numerous citrullinated microbial and food proteins.Arthritis Rheumatol. 2015 May;67(8):2020-31. doi: 10.1002/art.39161. Arthritis Rheumatol. 2015. PMID: 25892475
-
Validation of a multiplex chip-based assay for the detection of autoantibodies against citrullinated peptides.Arthritis Res Ther. 2012 Oct 1;14(5):R201. doi: 10.1186/ar4039. Arthritis Res Ther. 2012. PMID: 23025688 Free PMC article.
-
Antibodies to several citrullinated antigens are enriched in the joints of rheumatoid arthritis patients.Arthritis Rheum. 2010 Jan;62(1):44-52. doi: 10.1002/art.25036. Arthritis Rheum. 2010. PMID: 20039432
-
Anti-citrullinated protein antibodies and their clinical utility in rheumatoid arthritis.Int J Rheum Dis. 2013 Aug;16(4):379-86. doi: 10.1111/1756-185X.12129. Epub 2013 Jul 15. Int J Rheum Dis. 2013. PMID: 23992255 Review.
-
Anti-citrullinated peptides as autoantigens in rheumatoid arthritis-relevance to treatment.Autoimmun Rev. 2014 Nov;13(11):1114-20. doi: 10.1016/j.autrev.2014.08.012. Epub 2014 Aug 23. Autoimmun Rev. 2014. PMID: 25182207 Review.
Cited by
-
Multiplexed proteomic biosensor platform for label-free real-time simultaneous kinetic screening of thousands of protein interactions.Commun Biol. 2025 Mar 21;8(1):468. doi: 10.1038/s42003-025-07844-z. Commun Biol. 2025. PMID: 40119113 Free PMC article.
-
AAgAtlas 1.0: a human autoantigen database.Nucleic Acids Res. 2017 Jan 4;45(D1):D769-D776. doi: 10.1093/nar/gkw946. Epub 2016 Oct 19. Nucleic Acids Res. 2017. PMID: 27924021 Free PMC article.
-
Insulin-Like Growth Factor Binding Protein 6 in Rheumatoid Arthritis: A Possible Novel Chemotactic Factor?Front Immunol. 2017 May 18;8:554. doi: 10.3389/fimmu.2017.00554. eCollection 2017. Front Immunol. 2017. PMID: 28572803 Free PMC article.
-
Identification of anti-citrullinated osteopontin antibodies and increased inflammatory response by enhancement of osteopontin binding to fibroblast-like synoviocytes in rheumatoid arthritis.Arthritis Res Ther. 2023 Feb 17;25(1):25. doi: 10.1186/s13075-023-03007-9. Arthritis Res Ther. 2023. PMID: 36804906 Free PMC article.
-
Breaking tolerance: autoantibodies can target protein posttranslational modifications.Curr Opin Biotechnol. 2024 Feb;85:103056. doi: 10.1016/j.copbio.2023.103056. Epub 2023 Dec 22. Curr Opin Biotechnol. 2024. PMID: 38141322 Free PMC article. Review.
References
-
- Zaenker P., and Ziman M. R. (2013) Serologic autoantibodies as diagnostic cancer biomarkers–a review. Cancer Epidemiol. Biomarkers Prevention 22, 2161–2181 - PubMed
-
- Anderton S. M. (2004) Post-translational modifications of self antigens: implications for autoimmunity. Curr. Opinion Immunol. 16, 753–758 - PubMed
-
- Cantaert T., De Rycke L., Bongartz T., Matteson E. L., Tak P. P., Nicholas A. P., and Baeten D. (2006) Citrullinated proteins in rheumatoid arthritis: crucial.but not sufficient! Arthritis Rheumatism 54, 3381–3389 - PubMed
-
- Brahms H., Raymackers J., Union A., de Keyser F., Meheus L., and Luhrmann R. (2000) The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J. Biol. Chem. 275, 17122–17129 - PubMed
-
- Dieterich W., Ehnis T., Bauer M., Donner P., Volta U., Riecken E. O., and Schuppan D. (1997) Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat. Med. 3, 797–801 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

