Characterizing the functions of Ty1 Gag and the Gag-derived restriction factor p22/p18
- PMID: 27141325
- PMCID: PMC4836483
- DOI: 10.1080/2159256X.2016.1154637
Characterizing the functions of Ty1 Gag and the Gag-derived restriction factor p22/p18
Abstract
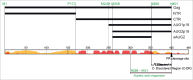
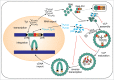
The long terminal repeat (LTR) and non-LTR retrotransposons comprise approximately half of the human genome, and we are only beginning to understand their influence on genome function and evolution. The LTR retrotransposon Ty1 is the most abundant mobile genetic element in the S. cerevisiae reference genome. Ty1 replicates via an RNA intermediate and shares several important structural and functional characteristics with retroviruses. However, unlike retroviruses Ty1 retrotransposition is not infectious. Retrotransposons integrations can cause mutations and genome instability. Despite the fact that S. cerevisiae lacks eukaryotic defense mechanisms such as RNAi, they maintain a relatively low copy number of the Ty1 retrotransposon in their genomes. A novel restriction factor derived from the C-terminal half of Gag (p22/p18) and encoded by internally initiated transcript inhibits retrotransposition in a dose-dependent manner. Therefore, Ty1 evolved a specific GAG organization and expression strategy to produce products both essential and antagonistic for retrotransposon movement. In this commentary we discuss our recent research aimed at defining steps of Ty1 replication influenced by p22/p18 with particular emphasis on the nucleic acid chaperone functions carried out by Gag and the restriction factor.
Keywords: Gag; RNA structure; Ty1 RNA; Ty1 retrotransposon; nucleic acid chaperone; restriction factor.
Figures
Comment on
- doi: 10.1093/nar/gkv695
Similar articles
-
A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control.J Virol. 2015 Apr;89(7):3922-38. doi: 10.1128/JVI.03060-14. Epub 2015 Jan 21. J Virol. 2015. PMID: 25609815 Free PMC article.
-
Ty1 escapes restriction by the self-encoded factor p22 through mutations in capsid.Mob Genet Elements. 2016 Mar 7;6(2):e1154639. doi: 10.1080/2159256X.2016.1154639. eCollection 2016 Mar-Apr. Mob Genet Elements. 2016. PMID: 27141327 Free PMC article.
-
Structure of Ty1 Internally Initiated RNA Influences Restriction Factor Expression.Viruses. 2017 Apr 10;9(4):74. doi: 10.3390/v9040074. Viruses. 2017. PMID: 28394277 Free PMC article.
-
A self-encoded capsid derivative restricts Ty1 retrotransposition in Saccharomyces.Curr Genet. 2016 May;62(2):321-9. doi: 10.1007/s00294-015-0550-6. Epub 2015 Dec 9. Curr Genet. 2016. PMID: 26650614 Free PMC article. Review.
-
The Ty1 LTR-Retrotransposon of Budding Yeast, Saccharomyces cerevisiae.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0053-2014. doi: 10.1128/microbiolspec.MDNA3-0053-2014. Microbiol Spectr. 2015. PMID: 26104690 Review.
Cited by
-
Elimination of virus-like particles reduces protein aggregation and extends replicative lifespan in Saccharomyces cerevisiae.Proc Natl Acad Sci U S A. 2024 Apr 2;121(14):e2313538121. doi: 10.1073/pnas.2313538121. Epub 2024 Mar 25. Proc Natl Acad Sci U S A. 2024. PMID: 38527193 Free PMC article.
-
Retrotransposon targeting to RNA polymerase III-transcribed genes.Mob DNA. 2018 Apr 23;9:14. doi: 10.1186/s13100-018-0119-2. eCollection 2018. Mob DNA. 2018. PMID: 29713390 Free PMC article. Review.
References
-
- Belcourt MF, Farabaugh PJ. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell 1990; 62:339-52; PMID:2164889; http://dx.doi.org/10.1016/0092-8674(90)90371-K - DOI - PMC - PubMed
-
- Malagon F, Jensen TH. The T body, a new cytoplasmic RNA granule in Saccharomyces cerevisiae. Mol Cell Biol 2008; 28:6022-32; PMID:18678648; http://dx.doi.org/10.1128/MCB.00684-08 - DOI - PMC - PubMed
-
- Malagon F, Jensen TH. T-body formation precedes virus-like particle maturation in S. cerevisiae. RNA Biol 2011; 8:184-9; PMID:21358276; http://dx.doi.org/10.4161/rna.8.2.14822 - DOI - PMC - PubMed
-
- Feng YX, Moore SP, Garfinkel DJ, Rein A. The genomic RNA in Ty1 virus-like particles is dimeric. J Virol 2000; 74:10819-21; PMID:11044130; http://dx.doi.org/10.1128/JVI.74.22.10819-10821.2000 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases