Helitrons in Drosophila: Chromatin modulation and tandem insertions
- PMID: 27141326
- PMCID: PMC4836490
- DOI: 10.1080/2159256X.2016.1154638
Helitrons in Drosophila: Chromatin modulation and tandem insertions
Abstract
Although Helitrons were discovered 15 y ago, they still represent an elusive group of transposable elements (TEs). They are thought to transpose via a rolling-circle mechanism, but no transposition assay has yet been conducted. We have recently characterized a group of Helitrons in Drosophila, named DINE-TR1, that display interesting features, including pronounced enrichment at β-heterochromatin, multiple tandem insertions (TIs) of the entire TE, and that experienced at least 2 independent expansion events of its internal tandem repeats (TRs) in distant Drosophila lineages. Here we discuss 2 aspects of TE dynamics displayed by the DINE-TR1 Helitrons: (i) the general evolutionary impact of piRNA-guided heterochromatin formation via TE-derived TR expansion and (ii) the possible mechanisms that could account for the recurrent TIs of Helitrons.
Keywords: DINE-1; chromatin sink; gene regulation; piRNA; β-heterochromatin.
Figures
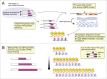
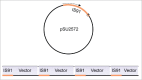

Comment on
- doi: 10.1007/s10577-015-9480-x
References
-
- Kidwell MG, Lisch DR. Transposable elements and host genome evolution. Trends Ecol Evol 2000; 15(3):95-9; PMID:10675923; http://dx.doi.org/ 10.1016/S0169-5347(99)01817-0 - DOI - PubMed
-
- Shapiro JA, von Sternberg R. Why repetitive DNA is essential to genome function. Biol Rev 2005; 80(02):227-50; PMID:15921050; http://dx.doi.org/ 10.1017/S1464793104006657 - DOI - PubMed
-
- Elliott TA, Linquist S, Gregory TR. Conceptual and empirical challenges of ascribing functions to transposable elements. Am Nat 2014; 184(1):14-24; PMID:24921597; http://dx.doi.org/ 10.1086/676588 - DOI - PubMed
-
- Gregory TR. Synergy between sequence and size in large-scale genomics. Nat Rev Genet 2005; 6(9):699-708; PMID:16151375; http://dx.doi.org/ 10.1038/nrg1674 - DOI - PubMed
-
- Kapitonov VV, Jurka J. Rolling-circle transposons in eukaryotes. Proc Natl Acad Sci USA 2001; 98(15):8714-9; http://dx.doi.org/ 10.1073/pnas.151269298 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
