Double-stranded RNA analog and type I interferon regulate expression of Trem paired receptors in murine myeloid cells
- PMID: 27141827
- PMCID: PMC4855714
- DOI: 10.1186/s12865-016-0147-y
Double-stranded RNA analog and type I interferon regulate expression of Trem paired receptors in murine myeloid cells
Abstract
Background: Triggering receptors expressed on myeloid cells (Trem) proteins are a family of cell surface receptors used to control innate immune responses such as proinflammatory cytokine production in mice. Trem genes belong to a rapidly expanding family of receptors that include activating and inhibitory paired-isoforms.
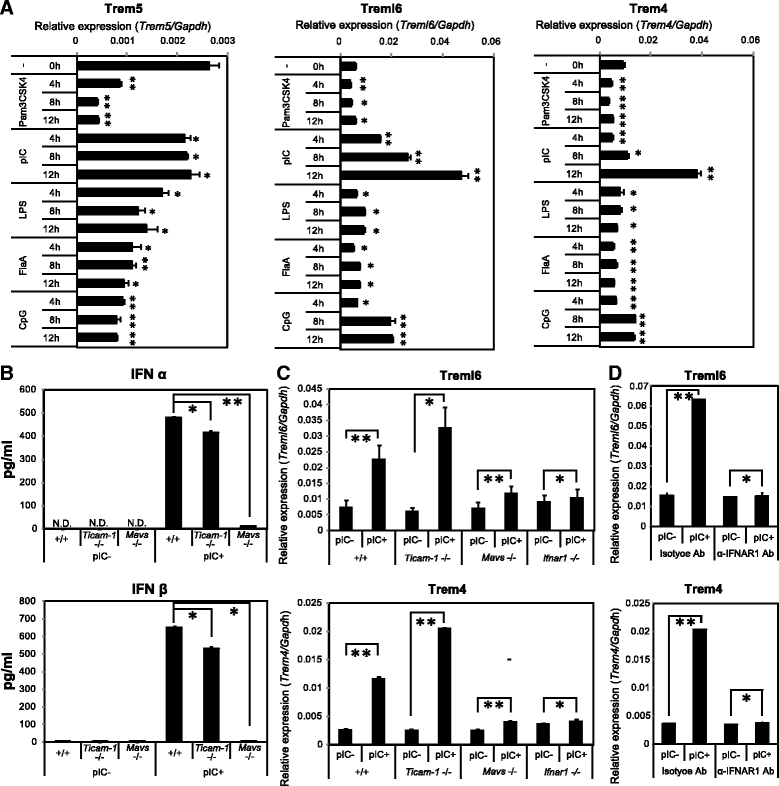
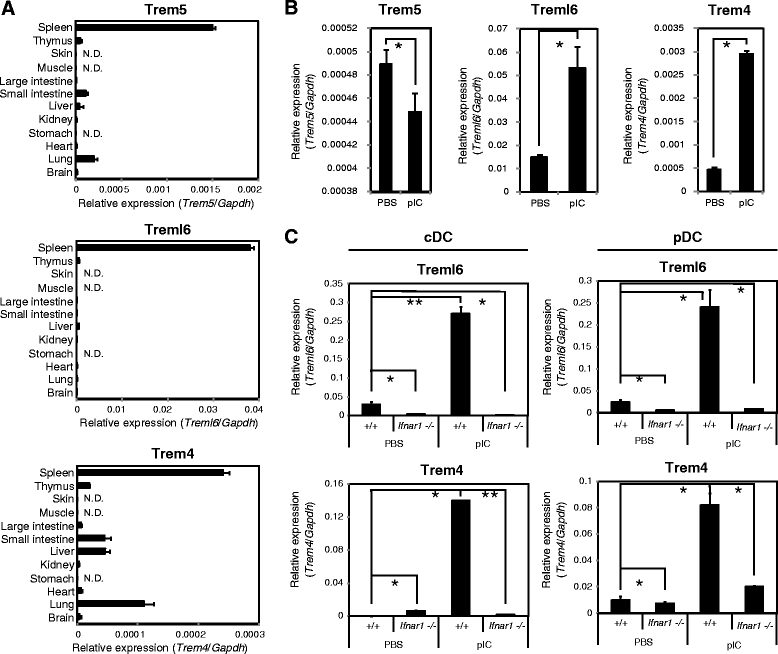
Results: By comparative genomic analysis, we found that Trem4, Trem5 and Trem-like transcript-6 (Treml6) genes typically paired receptors. These paired Trem genes were murine-specific and originated from an immunoreceptor tyrosine-based inhibition motif (ITIM)-containing gene. Treml6 encoded ITIM, whereas Trem4 and Trem5 lacked the ITIM but possessed positively-charged residues to associate with DNAX activating protein of 12 kDa (DAP12). DAP12 was directly associated with Trem4 and Trem5, and DAP12 coupling was mandatory for their expression on the cell surface. In bone marrow-derived dendritic cells (BMDCs) and macrophages (BMDMs), and splenic DC subsets, polyinosinic-polycytidylic acid (polyI:C) followed by type I interferon (IFN) production induced Trem4 and Treml6 whereas polyI:C or other TLR agonists failed to induce the expression of Trem5. PolyI:C induced Treml6 and Trem4 more efficiently in BMDMs than BMDCs. Treml6 was more potentially up-regulated in conventional DC (cDCs) and plasmacytoid DC (pDCs) than Trem4 in mice upon in vivo stimulation with polyI:C.
Discussion: Treml6-dependent inhibitory signal would be dominant in viral infection compared to resting state. Though no direct ligands of these Trem receptors have been determined, the results infer that a set of Trem receptors are up-regulated in response to viral RNA to regulate myeloid cell activation through modulation of DAP12-associated Trem4 and ITIM-containing Treml6.
Keywords: Dendritic cells; Evolution; Macrophages; Paired receptors; RNA sensors; Trem family; Type I interferon.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

