Whole Genome Re-Sequencing Identifies a Quantitative Trait Locus Repressing Carbon Reserve Accumulation during Optimal Growth in Chlamydomonas reinhardtii
- PMID: 27141848
- PMCID: PMC4855234
- DOI: 10.1038/srep25209
Whole Genome Re-Sequencing Identifies a Quantitative Trait Locus Repressing Carbon Reserve Accumulation during Optimal Growth in Chlamydomonas reinhardtii
Abstract
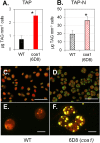
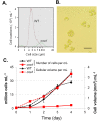
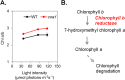
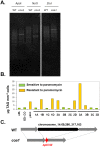
Microalgae have emerged as a promising source for biofuel production. Massive oil and starch accumulation in microalgae is possible, but occurs mostly when biomass growth is impaired. The molecular networks underlying the negative correlation between growth and reserve formation are not known. Thus isolation of strains capable of accumulating carbon reserves during optimal growth would be highly desirable. To this end, we screened an insertional mutant library of Chlamydomonas reinhardtii for alterations in oil content. A mutant accumulating five times more oil and twice more starch than wild-type during optimal growth was isolated and named constitutive oil accumulator 1 (coa1). Growth in photobioreactors under highly controlled conditions revealed that the increase in oil and starch content in coa1 was dependent on light intensity. Genetic analysis and DNA hybridization pointed to a single insertional event responsible for the phenotype. Whole genome re-sequencing identified in coa1 a >200 kb deletion on chromosome 14 containing 41 genes. This study demonstrates that, 1), the generation of algal strains accumulating higher reserve amount without compromising biomass accumulation is feasible; 2), light is an important parameter in phenotypic analysis; and 3), a chromosomal region (Quantitative Trait Locus) acts as suppressor of carbon reserve accumulation during optimal growth.
Figures


References
-
- Work V. H., D’Adamo S., Radakovits R., Jinkerson R. E. & Posewitz M. C. Improving photosynthesis and metabolic networks for the competitive production of phototroph-derived biofuels. Current Opinion in Biotechnology 23, 290–297 (2012). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

