Display of recombinant proteins at the surface of lactic acid bacteria: strategies and applications
- PMID: 27142045
- PMCID: PMC4855500
- DOI: 10.1186/s12934-016-0468-9
Display of recombinant proteins at the surface of lactic acid bacteria: strategies and applications
Abstract
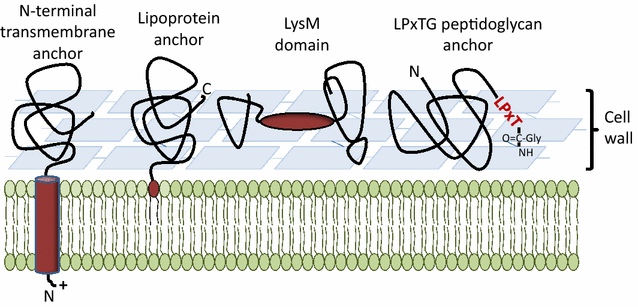
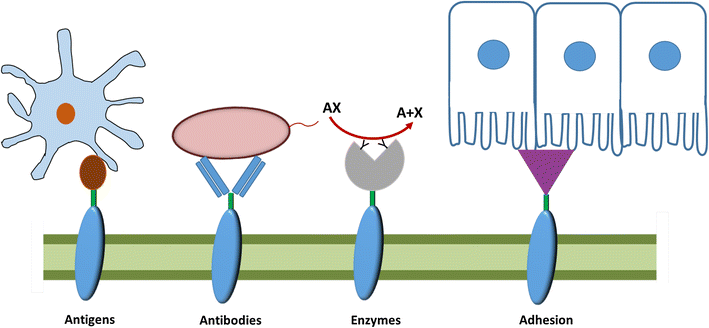
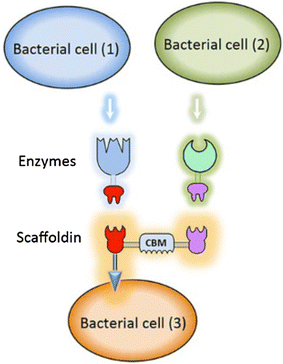
Lactic acid bacteria (LAB) are promising vectors of choice to deliver active molecules to mucosal tissues. They are recognized as safe by the World Health Organization and some strains have probiotic properties. The wide range of potential applications of LAB-driven mucosal delivery includes control of inflammatory bowel disease, vaccine delivery, and management of auto-immune diseases. Because of this potential, strategies for the display of proteins at the surface of LAB are gaining interest. To display a protein at the surface of LAB, a signal peptide and an anchor domain are necessary. The recombinant protein can be attached to the membrane layer, using a transmembrane anchor or a lipoprotein-anchor, or to the cell wall, by a covalent link using sortase mediated anchoring via the LPXTG motif, or by non-covalent liaisons employing binding domains such as LysM or WxL. Both the stability and functionality of the displayed proteins will be affected by the kind of anchor used. The most commonly surfaced exposed recombinant proteins produced in LAB are antigens and antibodies and the most commonly used LAB are lactococci and lactobacilli. Although it is not necessarily so that surface-display is the preferred localization in all cases, it has been shown that for certain applications, such as delivery of the human papillomavirus E7 antigen, surface-display elicits better biological responses, compared to cytosolic expression or secretion. Recent developments include the display of peptides and proteins targeting host cell receptors, for the purpose of enhancing the interactions between LAB and host. Surface-display technologies have other potential applications, such as degradation of biomass, which is of importance for some potential industrial applications of LAB.
Keywords: Genetic engineering; Lactic acid bacteria; Surface display.
Figures

References
-
- Motta J-P, Bermúdez-Humarán LG, Deraison C, Martin L, Rolland C, Rousset P, Boue J, Dietrich G, Chapman K, Kharrat P, Vinel J-P, Alric L, Mas E, Sallenave J-M, Langella P, Vergnolle N. Food-grade bacteria expressing elafin protect against inflammation and restore colon homeostasis. Sci Transl Med. 2012;4:158ra144. doi: 10.1126/scitranslmed.3004212. - DOI - PubMed
-
- Galipeau HJ, Wiepjes M, Motta J-P, Schulz JD, Jury J, Natividad JM, Pinto-Sanchez I, Sinclair D, Rousset P, Martin-Rosique R, Bermudez-Humaran L, Leroux JC, Murray JA, Smecuol E, Bai JC, Vergnolle N, Langella P, Verdu EF. Novel role of the serine protease inhibitor elafin in gluten-related disorders. Am J Gastroenterol. 2013;2014:1–9. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

