Single-cell profiling of human megakaryocyte-erythroid progenitors identifies distinct megakaryocyte and erythroid differentiation pathways
- PMID: 27142433
- PMCID: PMC4855892
- DOI: 10.1186/s13059-016-0939-7
Single-cell profiling of human megakaryocyte-erythroid progenitors identifies distinct megakaryocyte and erythroid differentiation pathways
Abstract
Background: Recent advances in single-cell techniques have provided the opportunity to finely dissect cellular heterogeneity within populations previously defined by "bulk" assays and to uncover rare cell types. In human hematopoiesis, megakaryocytes and erythroid cells differentiate from a shared precursor, the megakaryocyte-erythroid progenitor (MEP), which remains poorly defined.
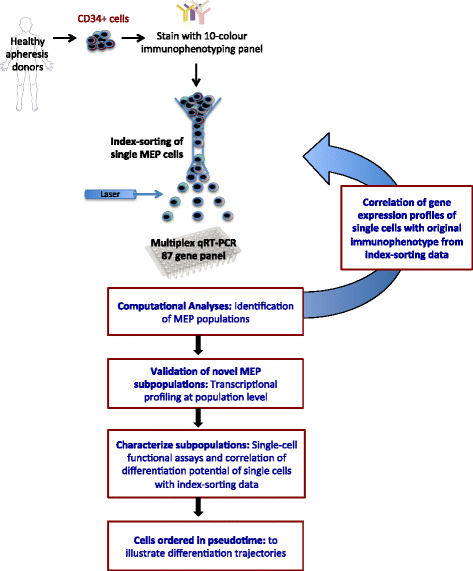
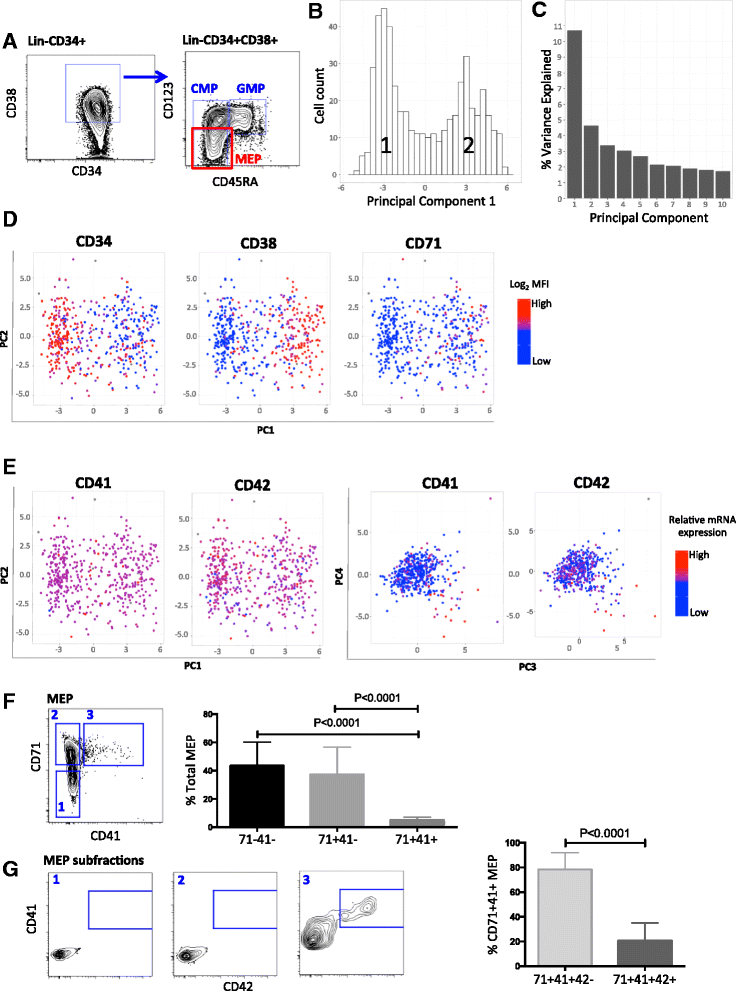
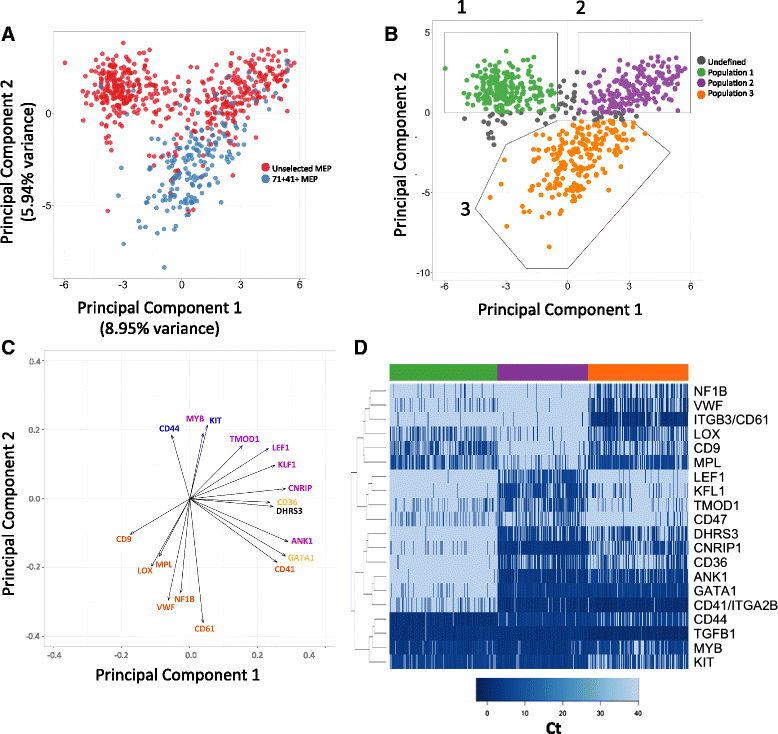
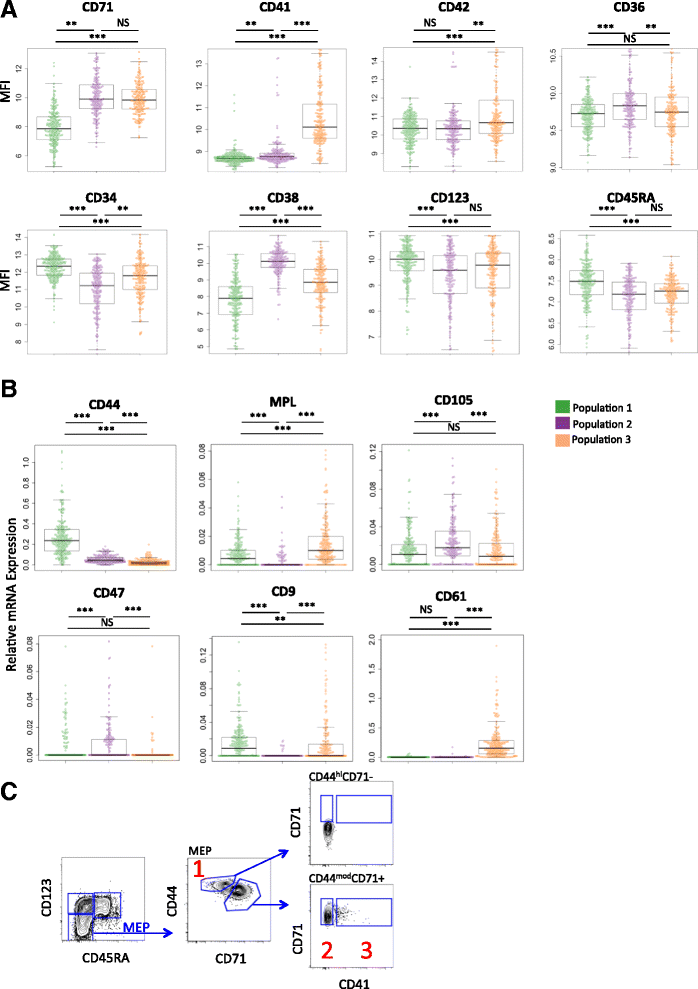
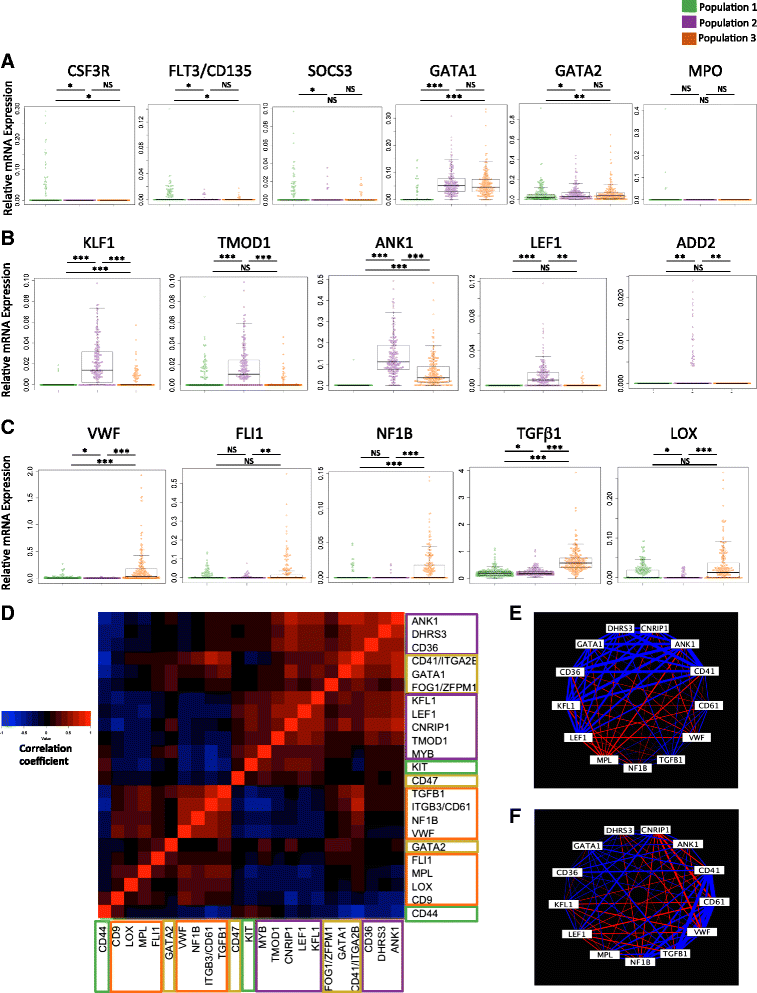
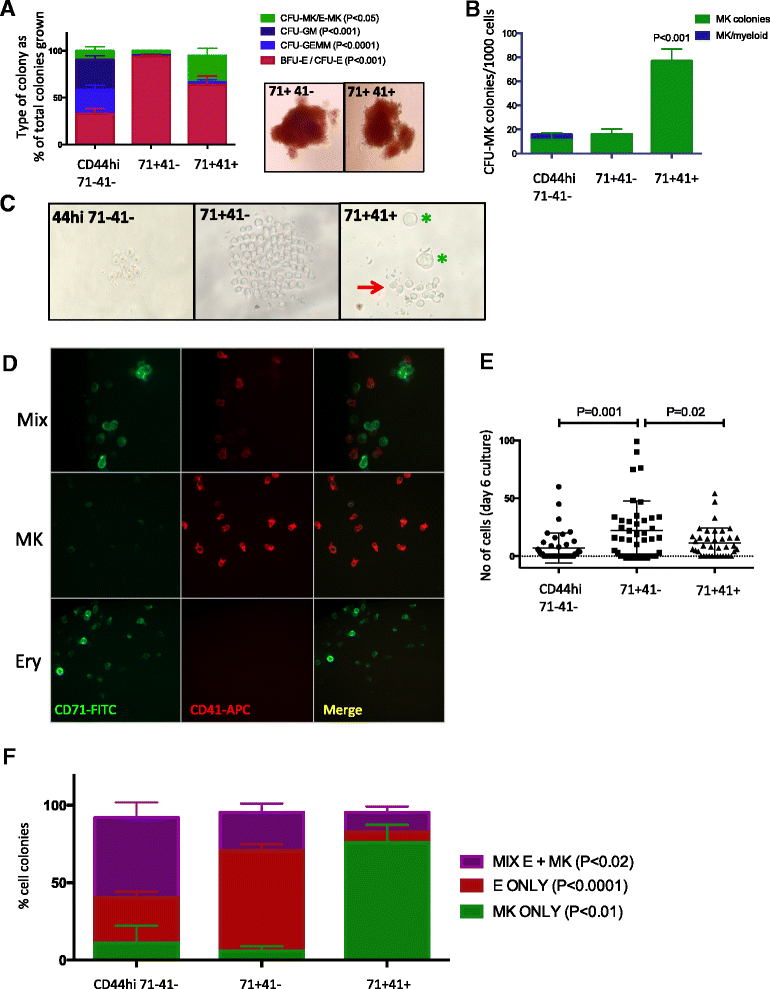
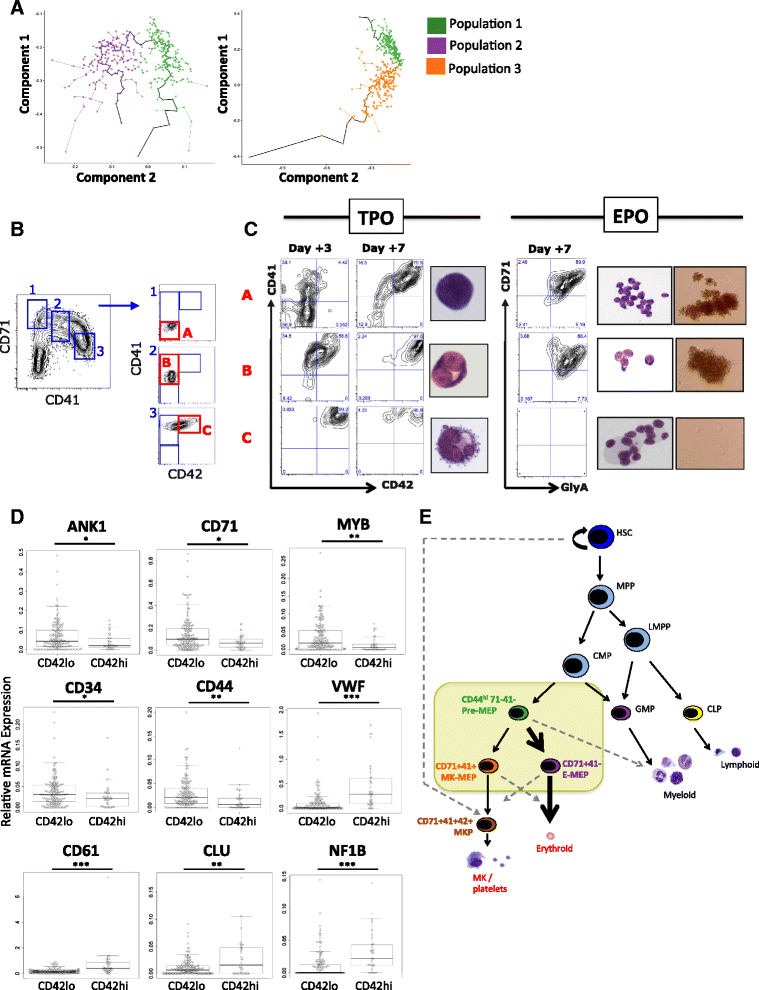
Results: To clarify the cellular pathway in erythro-megakaryocyte differentiation, we correlate the surface immunophenotype, transcriptional profile, and differentiation potential of individual MEP cells. Highly purified, single MEP cells were analyzed using index fluorescence-activated cell sorting and parallel targeted transcriptional profiling of the same cells was performed using a specifically designed panel of genes. Differentiation potential was tested in novel, single-cell differentiation assays. Our results demonstrate that immunophenotypic MEP comprise three distinct subpopulations: "Pre-MEP," enriched for erythroid/megakaryocyte progenitors but with residual myeloid differentiation capacity; "E-MEP," strongly biased towards erythroid differentiation; and "MK-MEP," a previously undescribed, rare population of cells that are bipotent but primarily generate megakaryocytic progeny. Therefore, conventionally defined MEP are a mixed population, as a minority give rise to mixed-lineage colonies while the majority of cells are transcriptionally primed to generate exclusively single-lineage output.
Conclusions: Our study clarifies the cellular hierarchy in human megakaryocyte/erythroid lineage commitment and highlights the importance of using a combination of single-cell approaches to dissect cellular heterogeneity and identify rare cell types within a population. We present a novel immunophenotyping strategy that enables the prospective identification of specific intermediate progenitor populations in erythro-megakaryopoiesis, allowing for in-depth study of disorders including inherited cytopenias, myeloproliferative disorders, and erythromegakaryocytic leukemias.
Keywords: Hematopoiesis; erythropoiesis; hematopoietic stem cell; megakaryopoiesis; myelopoiesis; thrombopoiesis.
Figures
References
-
- Debili N, Coulombel L, Croisille L, Katz A, Guichard J, Breton-Gorius J, et al. Characterization of a bipotent erythro-megakaryocytic progenitor in human bone marrow. Blood. 1996;88:1284–1296. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

