Utilization of HIV-1 envelope V3 to identify X4- and R5-specific Tat and LTR sequence signatures
- PMID: 27143130
- PMCID: PMC4855882
- DOI: 10.1186/s12977-016-0266-9
Utilization of HIV-1 envelope V3 to identify X4- and R5-specific Tat and LTR sequence signatures
Abstract
Background: HIV-1 entry is a receptor-mediated process directed by the interaction of the viral envelope with the host cell CD4 molecule and one of two co-receptors, CCR5 or CXCR4. The amino acid sequence of the third variable (V3) loop of the HIV-1 envelope is highly predictive of co-receptor utilization preference during entry, and machine learning predictive algorithms have been developed to characterize sequences as CCR5-utilizing (R5) or CXCR4-utilizing (X4). It was hypothesized that while the V3 loop is predominantly responsible for determining co-receptor binding, additional components of the HIV-1 genome may contribute to overall viral tropism and display sequence signatures associated with co-receptor utilization.
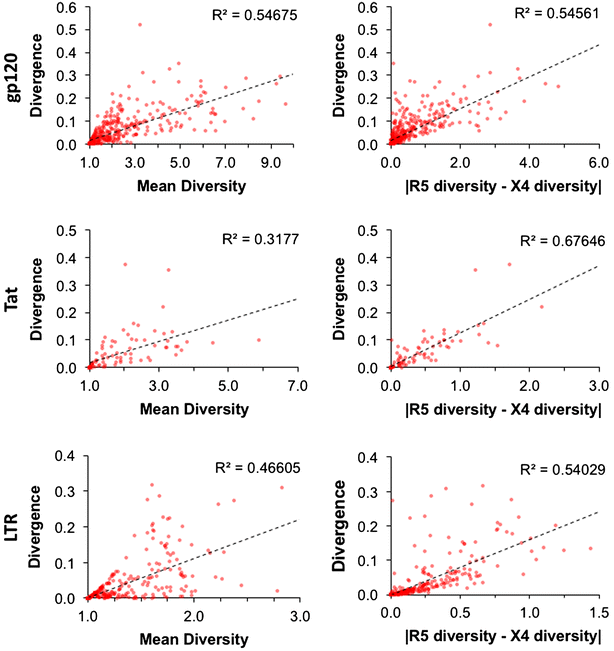
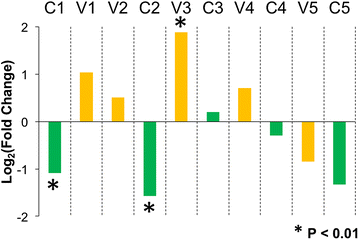
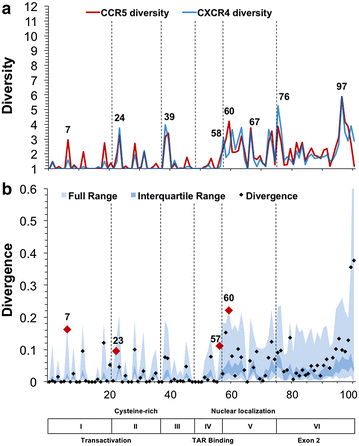
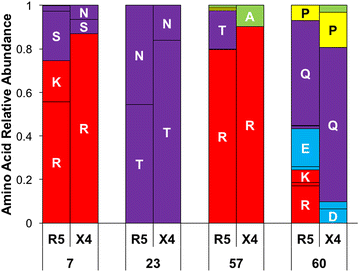
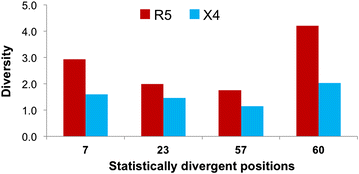
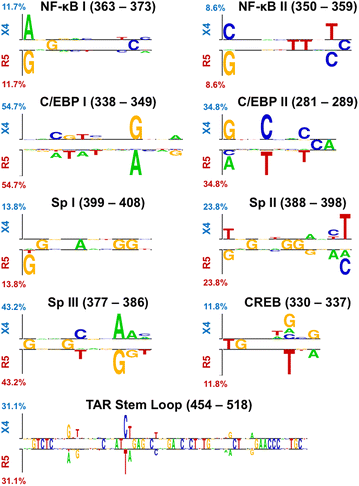
Results: The accessory protein Tat and the HlV-1 long terminal repeat (LTR) were analyzed with respect to genetic diversity and compared by Jensen-Shannon divergence which resulted in a correlation with both mean genetic diversity as well as the absolute difference in genetic diversity between R5- and X4-genome specific trends. As expected, the V3 domain of the gp120 protein was enriched with statistically divergent positions. Statistically divergent positions were also identified in Tat amino acid sequences within the transactivation and TAR-binding domains, and in nucleotide positions throughout the LTR. We further analyzed LTR sequences for putative transcription factor binding sites using the JASPAR transcription factor binding profile database and found several putative differences in transcription factor binding sites between R5 and X4 HIV-1 genomes, specifically identifying the C/EBP sites I and II, and Sp site III to differ with respect to sequence configuration for R5 and X4 LTRs.
Conclusion: These observations support the hypothesis that co-receptor utilization coincides with specific genetic signatures in HIV-1 Tat and the LTR, likely due to differing transcriptional regulatory mechanisms and selective pressures applied within specific cellular targets during the course of productive HIV-1 infection.
Keywords: Co-receptor; Divergence; Diversity; HIV-1; LTR; Tat; Transcription factor; Tropism; V3; gp120.
Figures



Similar articles
-
On the Physicochemical and Structural Modifications Associated with HIV-1 Subtype B Tropism Transition.AIDS Res Hum Retroviruses. 2016 Aug;32(8):829-40. doi: 10.1089/AID.2015.0373. Epub 2016 Jun 1. AIDS Res Hum Retroviruses. 2016. PMID: 27071630 Free PMC article.
-
Defining differential genetic signatures in CXCR4- and the CCR5-utilizing HIV-1 co-linear sequences.PLoS One. 2014 Sep 29;9(9):e107389. doi: 10.1371/journal.pone.0107389. eCollection 2014. PLoS One. 2014. PMID: 25265194 Free PMC article.
-
Selected amino acid mutations in HIV-1 B subtype gp41 are associated with specific gp120v₃ signatures in the regulation of co-receptor usage.Retrovirology. 2011 May 12;8:33. doi: 10.1186/1742-4690-8-33. Retrovirology. 2011. PMID: 21569409 Free PMC article.
-
HIV-1 subtype C predicted co-receptor tropism in Africa: an individual sequence level meta-analysis.AIDS Res Ther. 2020 Feb 7;17(1):5. doi: 10.1186/s12981-020-0263-x. AIDS Res Ther. 2020. PMID: 32033571 Free PMC article. Review.
-
Effect of HIV-1 subtype and tropism on treatment with chemokine coreceptor entry inhibitors; overview of viral entry inhibition.Crit Rev Microbiol. 2015;41(4):473-87. doi: 10.3109/1040841X.2013.867829. Epub 2014 Mar 17. Crit Rev Microbiol. 2015. PMID: 24635642 Review.
Cited by
-
Novel gRNA design pipeline to develop broad-spectrum CRISPR/Cas9 gRNAs for safe targeting of the HIV-1 quasispecies in patients.Sci Rep. 2019 Nov 19;9(1):17088. doi: 10.1038/s41598-019-52353-9. Sci Rep. 2019. PMID: 31745112 Free PMC article.
-
EcoHIV infection of mice establishes latent viral reservoirs in T cells and active viral reservoirs in macrophages that are sufficient for induction of neurocognitive impairment.PLoS Pathog. 2018 Jun 7;14(6):e1007061. doi: 10.1371/journal.ppat.1007061. eCollection 2018 Jun. PLoS Pathog. 2018. PMID: 29879225 Free PMC article.
-
Beyond Infection: The Role of Secreted Viral Proteins in Pathogenesis, Disease Severity and Diagnostic Applications.Cells. 2025 Apr 22;14(9):624. doi: 10.3390/cells14090624. Cells. 2025. PMID: 40358148 Free PMC article. Review.
-
Defining the molecular mechanisms of HIV-1 Tat secretion: PtdIns(4,5)P2 at the epicenter.Traffic. 2018 Apr 30:10.1111/tra.12578. doi: 10.1111/tra.12578. Online ahead of print. Traffic. 2018. PMID: 29708629 Free PMC article. Review.
-
Investigating the distribution of HIV-1 Tat lengths present in the Drexel Medicine CARES cohort.Virus Res. 2019 Oct 15;272:197727. doi: 10.1016/j.virusres.2019.197727. Epub 2019 Aug 19. Virus Res. 2019. PMID: 31437485 Free PMC article.
References
-
- Javaherian K, Langlois AJ, McDanal C, Ross KL, Eckler LI, Jellis CL, Profy AT, Rusche JR, Bolognesi DP, Putney SD, et al. Principal neutralizing domain of the human immunodeficiency virus type 1 envelope protein. Proc Natl Acad Sci USA. 1989;86:6768–6772. doi: 10.1073/pnas.86.17.6768. - DOI - PMC - PubMed
-
- Goudsmit J, Debouck C, Meloen RH, Smit L, Bakker M, Asher DM, Wolff AV, Gibbs CJ, Jr, Gajdusek DC. Human immunodeficiency virus type 1 neutralization epitope with conserved architecture elicits early type-specific antibodies in experimentally infected chimpanzees. Proc Natl Acad Sci USA. 1988;85:4478–4482. doi: 10.1073/pnas.85.12.4478. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

