Single-Cell Genome and Group-Specific dsrAB Sequencing Implicate Marine Members of the Class Dehalococcoidia (Phylum Chloroflexi) in Sulfur Cycling
- PMID: 27143384
- PMCID: PMC4959651
- DOI: 10.1128/mBio.00266-16
Single-Cell Genome and Group-Specific dsrAB Sequencing Implicate Marine Members of the Class Dehalococcoidia (Phylum Chloroflexi) in Sulfur Cycling
Abstract
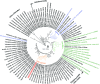
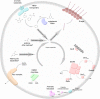
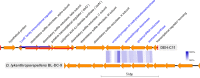
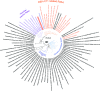
The marine subsurface sediment biosphere is widely inhabited by bacteria affiliated with the class Dehalococcoidia (DEH), phylum Chloroflexi, and yet little is known regarding their metabolisms. In this report, genomic content from a single DEH cell (DEH-C11) with a 16S rRNA gene that was affiliated with a diverse cluster of 16S rRNA gene sequences prevalent in marine sediments was obtained from sediments of Aarhus Bay, Denmark. The distinctive gene content of this cell suggests metabolic characteristics that differ from those of known DEH and Chloroflexi The presence of genes encoding dissimilatory sulfite reductase (Dsr) suggests that DEH could respire oxidized sulfur compounds, although Chloroflexi have never been implicated in this mode of sulfur cycling. Using long-range PCR assays targeting DEH dsr loci, dsrAB genes were amplified and sequenced from various marine sediments. Many of the amplified dsrAB sequences were affiliated with the DEH Dsr clade, which we propose equates to a family-level clade. This provides supporting evidence for the potential for sulfite reduction by diverse DEH species. DEH-C11 also harbored genes encoding reductases for arsenate, dimethyl sulfoxide, and halogenated organics. The reductive dehalogenase homolog (RdhA) forms a monophyletic clade along with RdhA sequences from various DEH-derived contigs retrieved from available metagenomes. Multiple facts indicate that this RdhA may not be a terminal reductase. The presence of other genes indicated that nutrients and energy may be derived from the oxidation of substituted homocyclic and heterocyclic aromatic compounds. Together, these results suggest that marine DEH play a previously unrecognized role in sulfur cycling and reveal the potential for expanded catabolic and respiratory functions among subsurface DEH.
Importance: Sediments underlying our oceans are inhabited by microorganisms in cell numbers similar to those estimated to inhabit the oceans. Microorganisms in sediments consist of various diverse and uncharacterized groups that contribute substantially to global biogeochemical cycles. Since most subsurface microorganisms continue to evade cultivation, possibly due to very slow growth, we obtained and analyzed genomic information from a representative of one of the most widespread and abundant, yet uncharacterized bacterial groups of the marine subsurface. We describe several key features that may contribute to their widespread distribution, such as respiratory flexibility and the potential to use oxidized sulfur compounds, which are abundant in marine environments, as electron acceptors. Together, these data provide important information that can be used to assist in designing enrichment strategies or other postgenomic studies, while also improving our understanding of the diversity and distribution of dsrAB genes, which are widely used functional marker genes for sulfur-cycling microbes.
Copyright © 2016 Wasmund et al.
Figures

Similar articles
-
Genome sequencing of a single cell of the widely distributed marine subsurface Dehalococcoidia, phylum Chloroflexi.ISME J. 2014 Feb;8(2):383-97. doi: 10.1038/ismej.2013.143. Epub 2013 Aug 22. ISME J. 2014. PMID: 23966099 Free PMC article.
-
Homoacetogenesis in Deep-Sea Chloroflexi, as Inferred by Single-Cell Genomics, Provides a Link to Reductive Dehalogenation in Terrestrial Dehalococcoidetes.mBio. 2017 Dec 19;8(6):e02022-17. doi: 10.1128/mBio.02022-17. mBio. 2017. PMID: 29259088 Free PMC article.
-
Comparative Single-Cell Genomics of Chloroflexi from the Okinawa Trough Deep-Subsurface Biosphere.Appl Environ Microbiol. 2016 May 2;82(10):3000-3008. doi: 10.1128/AEM.00624-16. Print 2016 May 15. Appl Environ Microbiol. 2016. PMID: 26969693 Free PMC article.
-
Development and application of primers for the class Dehalococcoidia (phylum Chloroflexi) enables deep insights into diversity and stratification of subgroups in the marine subsurface.Environ Microbiol. 2015 Oct;17(10):3540-56. doi: 10.1111/1462-2920.12510. Epub 2014 Jun 26. Environ Microbiol. 2015. PMID: 24889097
-
The life sulfuric: microbial ecology of sulfur cycling in marine sediments.Environ Microbiol Rep. 2017 Aug;9(4):323-344. doi: 10.1111/1758-2229.12538. Epub 2017 May 5. Environ Microbiol Rep. 2017. PMID: 28419734 Free PMC article. Review.
Cited by
-
Microbial community and geochemical analyses of trans-trench sediments for understanding the roles of hadal environments.ISME J. 2020 Mar;14(3):740-756. doi: 10.1038/s41396-019-0564-z. Epub 2019 Dec 11. ISME J. 2020. PMID: 31827245 Free PMC article.
-
Potential coupling of microbial methane, nitrogen, and sulphur cycling in the Okinawa Trough cold seep sediments.Microbiol Spectr. 2024 Jun 4;12(6):e0349023. doi: 10.1128/spectrum.03490-23. Epub 2024 May 1. Microbiol Spectr. 2024. PMID: 38690913 Free PMC article.
-
Roles of Organohalide-Respiring Dehalococcoidia in Carbon Cycling.mSystems. 2020 Jun 9;5(3):e00757-19. doi: 10.1128/mSystems.00757-19. mSystems. 2020. PMID: 32518199 Free PMC article.
-
SAR202 Genomes from the Dark Ocean Predict Pathways for the Oxidation of Recalcitrant Dissolved Organic Matter.mBio. 2017 Apr 18;8(2):e00413-17. doi: 10.1128/mBio.00413-17. mBio. 2017. PMID: 28420738 Free PMC article.
-
Novel Microorganisms Contribute to Biosulfidogenesis in the Deep Layer of an Acidic Pit Lake.Front Bioeng Biotechnol. 2022 Jul 13;10:867321. doi: 10.3389/fbioe.2022.867321. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35910036 Free PMC article.
References
-
- D’Hondt S, Jørgensen BB, Miller DJ, Batzke A, Blake R, Cragg BA, Cypionka H, Dickens GR, Ferdelman T, Hinrichs KU, Holm NG, Mitterer R, Spivack A, Wang G, Bekins B, Engelen B, Ford K, Gettemy G, Rutherford SD, Sass H, Skilbeck CG, Aiello IW, Guerin G, House CH, Inagaki F, Meister P, Naehr T, Niitsuma S, Parkes RJ, Schippers A, Smith DC, Teske A, Wiegel J, Padilla CN, Acosta JL. 2004. Distributions of microbial activities in deep subseafloor sediments. Science 306:2216–2221. doi: 10.1126/science.1101155. - DOI - PubMed
-
- Parkes RJ, Cragg B, Roussel E, Webster G, Weightman A, Sass H. 2014. A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere:geosphere interactions. Mar Geol 352:409–425. doi: 10.1016/j.margeo.2014.02.009. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

