Glycolipid GD3 and GD3 synthase are key drivers for glioblastoma stem cells and tumorigenicity
- PMID: 27143722
- PMCID: PMC4878508
- DOI: 10.1073/pnas.1604721113
Glycolipid GD3 and GD3 synthase are key drivers for glioblastoma stem cells and tumorigenicity
Abstract
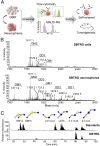
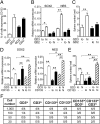
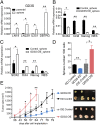
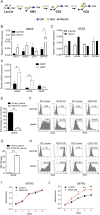
The cancer stem cells (CSCs) of glioblastoma multiforme (GBM), a grade IV astrocytoma, have been enriched by the expressed marker CD133. However, recent studies have shown that CD133(-) cells also possess tumor-initiating potential. By analysis of gangliosides on various cells, we show that ganglioside D3 (GD3) is overexpressed on eight neurospheres and tumor cells; in combination with CD133, the sorted cells exhibit a higher expression of stemness genes and self-renewal potential; and as few as six cells will form neurospheres and 20-30 cells will grow tumor in mice. Furthermore, GD3 synthase (GD3S) is increased in neurospheres and human GBM tissues, but not in normal brain tissues, and suppression of GD3S results in decreased GBM stem cell (GSC)-associated properties. In addition, a GD3 antibody is shown to induce complement-dependent cytotoxicity against cells expressing GD3 and inhibition of GBM tumor growth in vivo. Our results demonstrate that GD3 and GD3S are highly expressed in GSCs, play a key role in glioblastoma tumorigenicity, and are potential therapeutic targets against GBM.
Keywords: GBM; ST8SIA1; cancer stem cells; gangliosides; glycosphingolipids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Sox2, a stemness gene, regulates tumor-initiating and drug-resistant properties in CD133-positive glioblastoma stem cells.J Chin Med Assoc. 2016 Oct;79(10):538-45. doi: 10.1016/j.jcma.2016.03.010. Epub 2016 Aug 13. J Chin Med Assoc. 2016. PMID: 27530866
-
Engagement of cellular prion protein with the co-chaperone Hsp70/90 organizing protein regulates the proliferation of glioblastoma stem-like cells.Stem Cell Res Ther. 2017 Apr 17;8(1):76. doi: 10.1186/s13287-017-0518-1. Stem Cell Res Ther. 2017. PMID: 28412969 Free PMC article.
-
Progenitor/Stem Cell Markers in Brain Adjacent to Glioblastoma: GD3 Ganglioside and NG2 Proteoglycan Expression.J Neuropathol Exp Neurol. 2016 Feb;75(2):134-47. doi: 10.1093/jnen/nlv012. J Neuropathol Exp Neurol. 2016. PMID: 26792897
-
Glycosphingolipids in human embryonic stem cells and breast cancer stem cells, and potential cancer therapy strategies based on their structures and functions.Glycoconj J. 2022 Apr;39(2):177-195. doi: 10.1007/s10719-021-10032-w. Epub 2022 Mar 10. Glycoconj J. 2022. PMID: 35267131 Review.
-
Glioblastoma and stem cells.Neoplasma. 2008;55(5):369-74. Neoplasma. 2008. PMID: 18665745 Review.
Cited by
-
St8sia1-deficiency in mice alters tumor environments of gliomas, leading to reduced disease severity.Nagoya J Med Sci. 2021 Aug;83(3):535-549. doi: 10.18999/nagjms.83.3.535. Nagoya J Med Sci. 2021. PMID: 34552288 Free PMC article.
-
A modular chemoenzymatic cascade strategy for the structure-customized assembly of ganglioside analogs.Commun Chem. 2024 Jan 18;7(1):17. doi: 10.1038/s42004-024-01102-9. Commun Chem. 2024. PMID: 38238524 Free PMC article.
-
GD3 ganglioside is a promising therapeutic target for glioma patients.Neurooncol Adv. 2024 Mar 19;6(1):vdae038. doi: 10.1093/noajnl/vdae038. eCollection 2024 Jan-Dec. Neurooncol Adv. 2024. PMID: 38590763 Free PMC article. Review.
-
Overexpression of ceramide synthase 1 increases C18-ceramide and leads to lethal autophagy in human glioma.Oncotarget. 2017 Oct 23;8(61):104022-104036. doi: 10.18632/oncotarget.21955. eCollection 2017 Nov 28. Oncotarget. 2017. PMID: 29262618 Free PMC article.
-
Unique challenges for glioblastoma immunotherapy-discussions across neuro-oncology and non-neuro-oncology experts in cancer immunology. Meeting Report from the 2019 SNO Immuno-Oncology Think Tank.Neuro Oncol. 2021 Mar 25;23(3):356-375. doi: 10.1093/neuonc/noaa277. Neuro Oncol. 2021. PMID: 33367885 Free PMC article. Review.
References
-
- Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. - PubMed
-
- Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353(8):811–822. - PubMed
-
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. - PubMed
-
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8(10):755–768. - PubMed
-
- Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

