Bacteriophage MS2 genomic RNA encodes an assembly instruction manual for its capsid
- PMID: 27144089
- PMCID: PMC4836477
- DOI: 10.1080/21597081.2016.1157666
Bacteriophage MS2 genomic RNA encodes an assembly instruction manual for its capsid
Erratum in
- doi: 10.1016/j.jmb.2015.11.014
Abstract
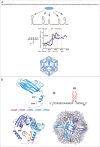
Using RNA-coat protein crosslinking we have shown that the principal RNA recognition surface on the interior of infectious MS2 virions overlaps with the known peptides that bind the high affinity translational operator, TR, within the phage genome. The data also reveal the sequences of genomic fragments in contact with the coat protein shell. These show remarkable overlap with previous predictions based on the hypothesis that virion assembly is mediated by multiple sequences-specific contacts at RNA sites termed Packaging Signals (PSs). These PSs are variations on the TR stem-loop sequence and secondary structure. They act co-operatively to regulate the dominant assembly pathway and ensure cognate RNA encapsidation. In MS2, they also trigger conformational change in the dimeric capsomere creating the A/B quasi-conformer, 60 of which are needed to complete the T=3 capsid. This is the most compelling demonstration to date that this ssRNA virus, and by implications potentially very many of them, assemble via a PS-mediated assembly mechanism.
Keywords: RNA bacteriophage; RNA packaging signals; virion assembly.
Figures

Similar articles
-
Viral genomic single-stranded RNA directs the pathway toward a T=3 capsid.J Mol Biol. 2010 Feb 5;395(5):924-36. doi: 10.1016/j.jmb.2009.11.018. Epub 2009 Nov 12. J Mol Biol. 2010. PMID: 19913556 Free PMC article.
-
Genome-regulated Assembly of a ssRNA Virus May Also Prepare It for Infection.J Mol Biol. 2022 Oct 30;434(20):167797. doi: 10.1016/j.jmb.2022.167797. Epub 2022 Aug 20. J Mol Biol. 2022. PMID: 35998704
-
Direct Evidence for Packaging Signal-Mediated Assembly of Bacteriophage MS2.J Mol Biol. 2016 Jan 29;428(2 Pt B):431-48. doi: 10.1016/j.jmb.2015.11.014. Epub 2015 Dec 1. J Mol Biol. 2016. PMID: 26608810 Free PMC article.
-
Protein-RNA Interactions in the Single-Stranded RNA Bacteriophages.Subcell Biochem. 2018;88:281-303. doi: 10.1007/978-981-10-8456-0_13. Subcell Biochem. 2018. PMID: 29900502 Review.
-
Molecular frustration: a hypothesis for regulation of viral infections.Trends Microbiol. 2024 Jan;32(1):17-26. doi: 10.1016/j.tim.2023.07.003. Epub 2023 Jul 27. Trends Microbiol. 2024. PMID: 37507296 Review.
Cited by
-
Rewriting nature's assembly manual for a ssRNA virus.Proc Natl Acad Sci U S A. 2017 Nov 14;114(46):12255-12260. doi: 10.1073/pnas.1706951114. Epub 2017 Oct 30. Proc Natl Acad Sci U S A. 2017. PMID: 29087310 Free PMC article.
-
Visualizing a viral genome with contrast variation small angle X-ray scattering.J Biol Chem. 2020 Nov 20;295(47):15923-15932. doi: 10.1074/jbc.RA120.013961. Epub 2020 Sep 10. J Biol Chem. 2020. PMID: 32913117 Free PMC article.
-
The Effect of RNA Secondary Structure on the Self-Assembly of Viral Capsids.Biophys J. 2017 Jul 25;113(2):339-347. doi: 10.1016/j.bpj.2017.06.038. Epub 2017 Jul 12. Biophys J. 2017. PMID: 28711172 Free PMC article.
-
Length of encapsidated cargo impacts stability and structure of in vitro assembled alphavirus core-like particles.J Phys Condens Matter. 2017 Dec 6;29(48):484003. doi: 10.1088/1361-648X/aa90d0. J Phys Condens Matter. 2017. PMID: 28975896 Free PMC article.
-
The importance of virion-incorporated cellular RNA-Binding Proteins in viral particle assembly and infectivity.Semin Cell Dev Biol. 2021 Mar;111:108-118. doi: 10.1016/j.semcdb.2020.08.002. Epub 2020 Sep 10. Semin Cell Dev Biol. 2021. PMID: 32921578 Free PMC article. Review.
References
-
- Schneemann A. The structural and functional role of RNA in icosahedral virus assembly. Annu Rev Microbiol 2006; 60:51-67; PMID:16704342; http://dx.doi.org/10.1146/annurev.micro.60.080805.142304 - DOI - PubMed
-
- van der Schoot P, Bruinsma R. Electrostatics and the assembly of an RNA virus. Phys Rev E Stat Nonlin Soft Matter Phys 2005; 71:061928; PMID:16089786; http://dx.doi.org/10.1103/PhysRevE.71.061928 - DOI - PubMed
-
- Belyi VA, Muthukumar M. Electrostatic origin of the genome packing in viruses. Proc Natl Acad Sci 2006;103:17174–8; PMID:17090672; http://dx.doi.org/406058710.1073/pnas.0608311103 - DOI - PMC - PubMed
-
- Balint R, Cohen SS. The incorporation of radiolabeled polyamines and methionine into turnip yellow mosaic virus in protoplasts from infected plants. Virology 1985; 144:181-93; PMID:4060587; http://dx.doi.org/10.1016/0042-6822(85)90316-2 - DOI - PubMed
-
- Bruinsma RF. Physics of RNA and viral assembly. Eur Phys J E Soft Matter 2006; 19:303-10; PMID:16554977; http://dx.doi.org/10.1140/epje/i2005-10071-1 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
