Oncogenic function of the homeobox A13-long noncoding RNA HOTTIP-insulin growth factor-binding protein 3 axis in human gastric cancer
- PMID: 27144338
- PMCID: PMC5094982
- DOI: 10.18632/oncotarget.9102
Oncogenic function of the homeobox A13-long noncoding RNA HOTTIP-insulin growth factor-binding protein 3 axis in human gastric cancer
Abstract
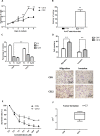
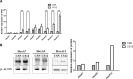
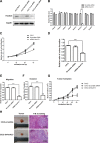
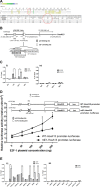
To study the mechanisms of gastric tumorigenesis, we have established CSN cell line from human normal gastric mucosa, and CS12, a tumorigenic and invasive gastric cancer cell line from CSN passages. Many stem cell markers were expressed in both CSN and CS12 cells, but LGR5 and NANOG were expressed only in CS12 cells. Increased expression of homeobox A13 (HoxA13) and its downstream cascades was significant for the tumorigenic activity of CS12 cells, and was associated with recruitment of E2F-1 to HoxA13 promoter accompanied with increased trimethylation of histone H3 lysine 4 (H3K4me3) at the hypomethylated E2F motifs. Knockdown of HoxA13 caused the downregulation of long non-coding RNA HOTTIP and insulin growth factor-binding protein 3 (IGFBP-3) genes, indicating that both were targets of HoxA13. Concurrent regulation of HoxA13-HOTTIP was mediated by the mixed lineage leukemia-WD repeat domain 5 complex, which caused the trimethylation of H3K4 and then stimulated cell proliferation. HoxA13 transactivated the IGFBP-3 promoter through the HOX-binding site. Activation of IGFBP-3 stimulated the oncogenic potential and invasion activity. Increased expression of HoxA13 (63.2%) and IGFBP-3 (28.6%) was detected in human gastric cancer tissues and was found in the gastric cancer data of The Cancer Genome Atlas. Taken together, the HoxA13-HOTTIP-IGFBP-3 cascade is critical for the carcinogenic characteristics of CS12 cells.
Keywords: HOTTIP; HoxA13; IGFBP-3; gastric cancer cells; p53-E2F signaling.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures


References
-
- Dolle P, Izpisua-Belmonte JC, Brown JM, Tickle C, Duboule D. HOX-4 genes and the morphogenesis of mammalian genitalia. Genes Dev. 1991;5:1767–1767. - PubMed
-
- Yokouchi Y, Sakiyama J, Kuroiwa A. Coordinated expression of Abd-B subfamily genes of the HoxA cluster in the developing digestive tract of chick embryo. Dev Biol. 1995;169:76–89. - PubMed
-
- Mortlock DP, Innis JW. Mutation of HOXA13 in hand-foot-genital syndrome. Nat Genet. 1997;15:179–180. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

